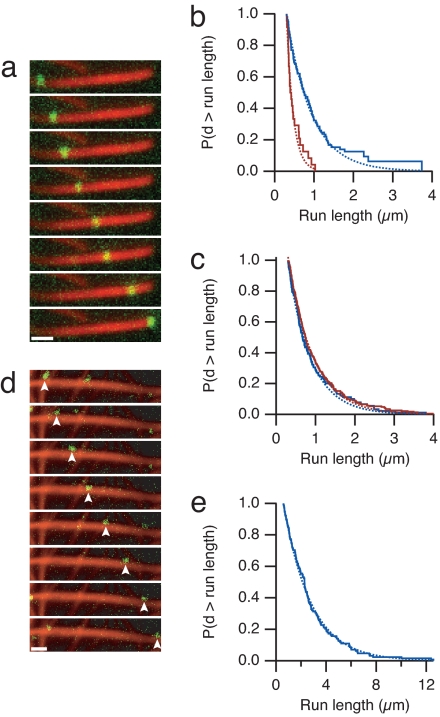
Fig. 2.
Processive motility of myosin X on actin bundles. (a) Time-lapse fluorescence micrographs of a single myosin X motor (green) moving along a fascin-actin bundle (red). (Frame interval, 2.5 s; scale bar, 1 μm.) (b) Run length measurements of myosin X on fascin-actin bundles (blue) and actin alone (red) at 2 mM ATP. The Kaplan–Meier estimate of the run-length survivor function is shown (38). Events are left-truncated at 0.3 μm and are right-censored at bundle ends. Run lengths are estimated from single exponential fits to the empirical survivor function (dotted lines). Run length decay constants are 0.63 ± 0.08 μm (SEM, n = 100) on fascin-actin bundles and 0.17 ± 0.05 μm (SEM, n = 24) on single actin filaments. This difference in run length is significant (P = 0.01, Kolmogorov–Smirnov test). Runs on single filaments were measured on 10-fold less actin that was observed for a 10-fold longer period, allowing us to directly compare the total number of events. Myosin X moved at 340 ± 120 nm/s (SD) on fascin-actin bundles, and 330 ± 120 nm/s (SD) on single filaments. (c) Run length measurements of myosin V on bundles and actin alone, as in b. Run length decay constants are 0.57 ± 0.06 μm (SEM, n = 134) on fascin-actin bundles, and 0.66 ± 0.05 μm (SEM, n = 231) on single actin filaments. Myosin V moved at 270 ± 110 nm/s (SD) on fascin-actin bundles, and 330 ± 100 nm/s (SD) on single filaments. (d) Time-lapse fluorescence micrographs of myosin X (green) moving along methylcellulose-bundled actin (red). Arrowheads indicate the moving spot. (Frame interval, 2.0 s; scale bar, 1 μm.) (e) The run length decay constant on methylcellulose-bundled actin is 2.6 ± 0.2 μm (SEM, n = 128). Myosin X moved at 780 ± 170 nm/s (SD) on methylcellulose-bundled actin. On some methylcellulose bundles, myosin X moved in a back-and-forth manner, suggesting that regions of the branched actin bundle network have mixed polarity. These mixed polarity bundles were excluded from the analysis. In control experiments, methylcellulose did not affect the run length of myosin X on fascin-actin bundles, and reduced the velocity by one-third. Run length standard errors are from fitting 200 bootstrap sampled sets. For single-molecule evidence, see Fig. S1.