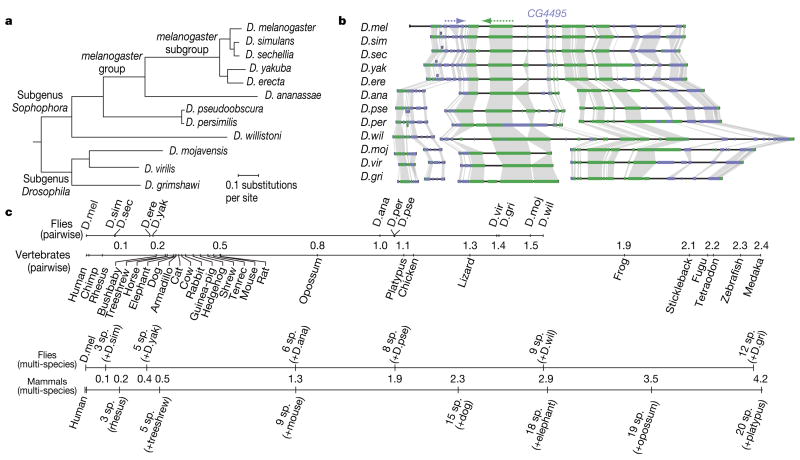
Figure 1. Phylogeny and alignment of 12 Drosophila species.
a, Phylogenetic tree relating the 12 Drosophila species, estimated from fourfold degenerate sites (Supplementary Methods 1). The 12 species span a total branch length of 4.13 substitutions per neutral site. b, Gene order conservation for a 0.45-Mb region of chromosome 2L centred on CG4495, for which we predict a new exon (Fig. 3a), and spanning 35 genes. Colour represents the direction of transcription. Boxes represent full gene models. Individual exons and introns are not shown. c, Comparison of evolutionary distances spanned by fly and vertebrate trees. Pairwise and multi-species distances (in substitutions per fourfold degenerate site) are shown from D. melanogaster and from human as reference genomes. Note that species with longer branches (for example, mouse) show higher pairwise distances, not always reflecting the order of divergence. Multi-species distances include all species within a phylogenetic clade.