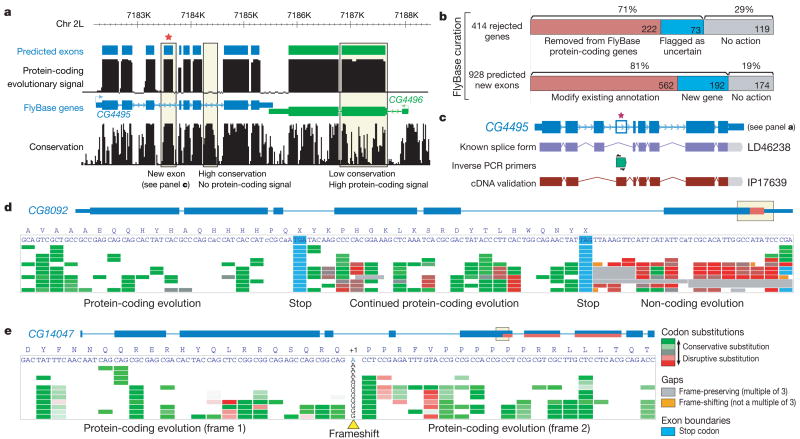
Figure 3. Revisiting the protein-coding gene catalogue and revealing unusual gene structures.
a, Protein-coding evolutionary signatures correlate with annotated protein-coding exons more precisely than the overall conservation level (phastCons track33), for example excluding highly conserved yet non-coding elements. Asterisk denotes new predicted exon, which we validate with cDNA sequencing (see panel c). The height of the black tracks indicates protein-coding potential according to evolutionary signatures (top) and overall sequence conservation (bottom). Blue and green boxes indicate predicted coding exons (top) and the current FlyBase annotation (bottom). The region shown represents the central 6 kb of Fig. 1b, rendered by the UCSC genome browser126. b, Results of FlyBase curation of 414 genes rejected by evolutionary signatures (Table 1), and 928 predicted new exons. c, Experimental validation of predicted new exon from panel a. Inverse PCR with primers in the predicted exon (green) results in a full-length cDNA clone, confirming the predicted exon and revealing a new alternative splice form for CG4495. d, Protein-coding evolution continues downstream of a conserved stop codon in 149 genes, suggesting translational readthrough. e, Codon-based evolutionary signatures (CSF score) abruptly shift from one reading frame to another within a protein-coding exon, suggesting a conserved, ‘programmed’ frameshift.