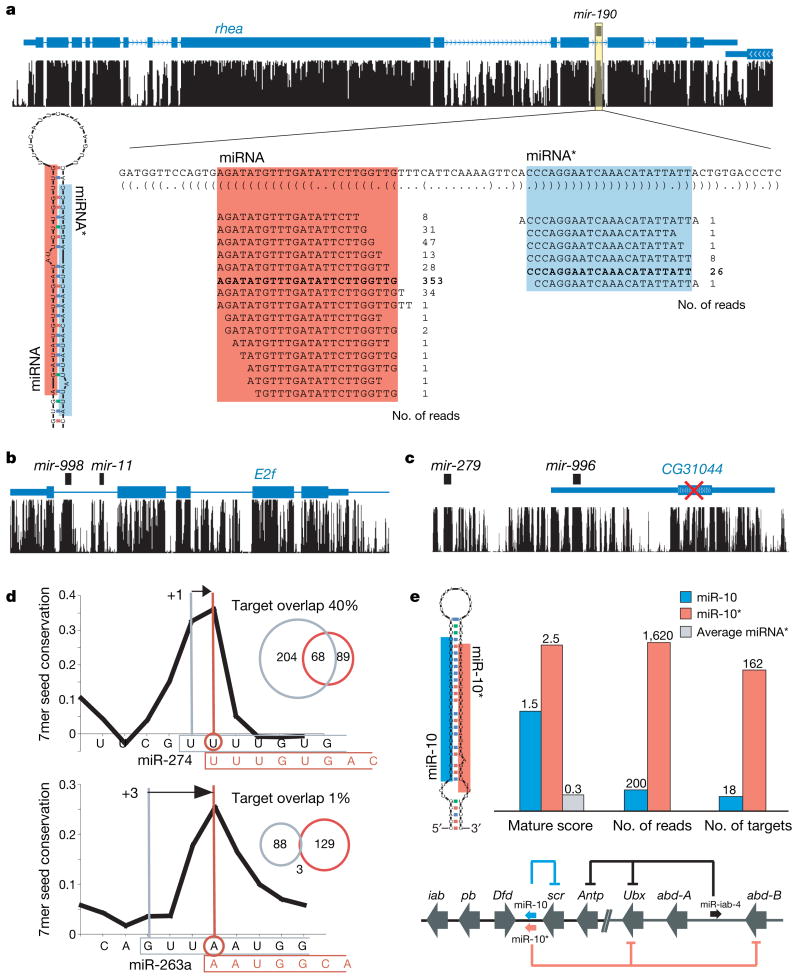
Figure 5. MicroRNA gene identification and functional implications.
a, New predicted miRNA (mir-190) and its validation by sequencing reads. Total read counts for mature miRNA (red) and miRNA* (blue) show a characteristic pattern of processing indicative of miRNAs. Highlighted regions indicate most abundant processing products. b, Example of clustered known (mir-11) and new (mir-998) miRNAs in the intron of cell-cycle regulator E2f. c, Example of a new miRNA (mir-996) in the transcript of a spurious gene. CG31044 was rejected by our protein-coding analysis, its transcript probably representing the precursor of mir-996, with no protein-coding function. d, Revisions to the 5′ end of miR-274 and miR-263a are proposed on the basis of evolutionary evidence (for example, 7mer seed conservation; black curve) and confirmed by sequencing reads. Changes at the 5′ end of more than one nucleotide results in marked changes to the predicted target spectra (venn diagrams). e, Evidence from evolutionary signals (mature score), sequencing reads and target predictions suggests that both miR-10 and miR-10* are functional, each targeting distinct Hox genes.