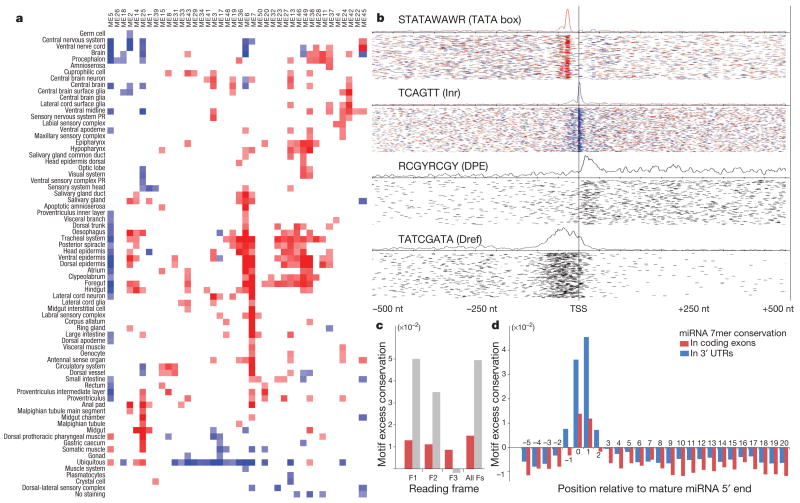
Figure 6. Regulatory motif discovery.
a, Discovered motifs show enrichment (red) or depletion (blue) in genes expressed in a given tissue (log colour range from P =10−5 enrichment to P =10−5 depletion). Bi-clustering reveals groups of motifs with similar tissue enrichment and groups of tissues with similar motif content. Full matrix and randomized control is shown in Supplementary Fig. 6d. b, Positional bias of discovered motifs relative to transcription start sites (TSS). Peaks with highly specific distances from the transcription start site (for example, first three plots) are characteristic of core promoter elements, and broad peaks (for example, fourth plot) are characteristic of transcription factors. For non-palindromic motifs, colours indicate forward-strand (red) and reverse-strand (blue) instances. Curves denote the density of all instances and individual segments denote individual motif instances, summed across groups of 50 genes (each line). c, Coding regions show reading-frame-invariant conservation for miRNA motifs (red) and reading-frame-biased conservation for protein motifs (grey). MEC scores are evaluated for each of the three reading frame offsets (F1–F3) and also without frame correction (all Fs). Plots show average MEC for all miRNA motifs and 500 top-scoring protein-coding motifs (based on MEC without frame correction). d, Motif excess conservation (MEC) of 7mer complements at different offsets with respect to miRNA 5′ end, averaged across all Rfam miRNAs. MEC scores evaluated in protein-coding regions and 3′ UTRs show a highly similar profile (correlation coefficient 0.96), suggesting similar evolutionary constraints.