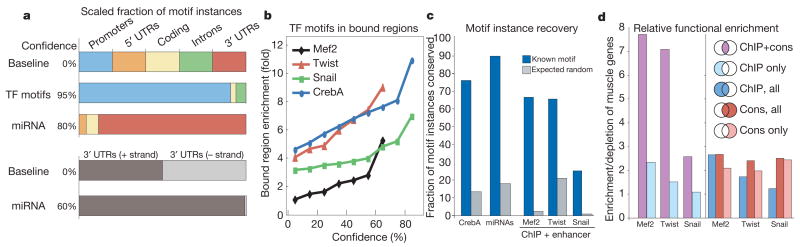
Figure 7. Identification of individual motif instances.
a, Increasing confidence levels select for motif instances in regions they are known to be functional: conserved transcription factor (TF) motifs enrich for promoters; miRNA motifs for 3′UTRs, and specifically the transcribed strand. Regions are normalized for their overall length, measured by the number of motif instances without conservation (0% confidence baseline). b, Increasing confidence levels select for transcription factor motif instances with experimental support for each factor tested. c, The high fraction of experimentally supported motif instances that are recovered at 60% confidence for transcription factors and 80% confidence for miRNAs illustrates the high sensitivity of the BLS approach. d, Comparison of chromatin immunoprecipitation (ChIP) and conservation in their ability to identify functional motif instances. Motif instances that are both ChIP-bound and conserved (purple) show the strongest functional enrichment in muscle genes for Mef2 and Twist (depletion for Snail), whereas motif instances derived by ChIP alone (light blue) show substantially reduced enrichment levels. Comparing the enrichment of all instances recovered by ChIP (blue) and all instances recovered by conservation (red) suggests that the two approaches perform comparably. Even the sites recovered by conservation alone outside bound regions (pink) show enrichment levels comparable to ChIP, suggesting that they are also functional.