Figure 3.
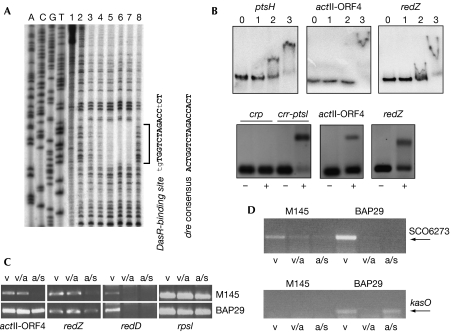
DasR controls antibiotic production in Streptomyces coelicolor. (A) Identification of the DasR-binding site by DNase I footprint analysis of the crr-ptsI upstream region. The crr-ptsI probe was incubated with DNase I (0.4 μg/ml) and increasing amounts of purified DasR (0, 10, 20, 40, 60 or 80 pmol of DasR in lanes 2, 3, 4, 5, 6 and 7, respectively). Controls: lane 1, no DasR or DNase I; lane 8, DNase I and 350 pmol of bovine serum albumin. ACGT, DNA sequence lanes. (B) Electrophoretic mobility gel shift assays showing direct interaction of DasR with dre sites predicted upstream from actII-ORF4 and redZ. Top: binding of DasR to the entire promoter regions of ptsH, actII-ORF4 and redZ; lane 0, probe; lanes 1, 2 and 3, probe and 1, 5 and 20 nM DasR, respectively; bottom: double-stranded oligonucleotide probes encompassing dre sites for crr-ptsI (positive control), actII-ORF4 and redZ were incubated with (+) or without (−) purified His-tagged DasR. The crp promoter region was used as a negative control. (C) Transcription of the pathway-specific activator genes for Act (actII-ORF4) and Red (redD) analysed by semiquantitative RT–PCR. Samples were collected from S. coelicolor M145 and the dasR mutant BAP29 grown on minimal medium mannitol plates after 30 h (vegetative growth (v)), 42 h (initiation of aerial growth (a)) and 72 h (aerial growth and spores (s)). rpsI (for ribosomal protein S9) was used as an RNA integrity control. (D) Transcriptional analysis of the kas ‘cryptic' type I polyketide cluster of S. coelicolor (SCO6273–6288) by semiquantitative RT–PCR. Transcriptional repression of kasO is relieved by deletion of dasR. RNA samples are as in (C). RT–PCR, reverse transcription–PCR.