Figure 2.
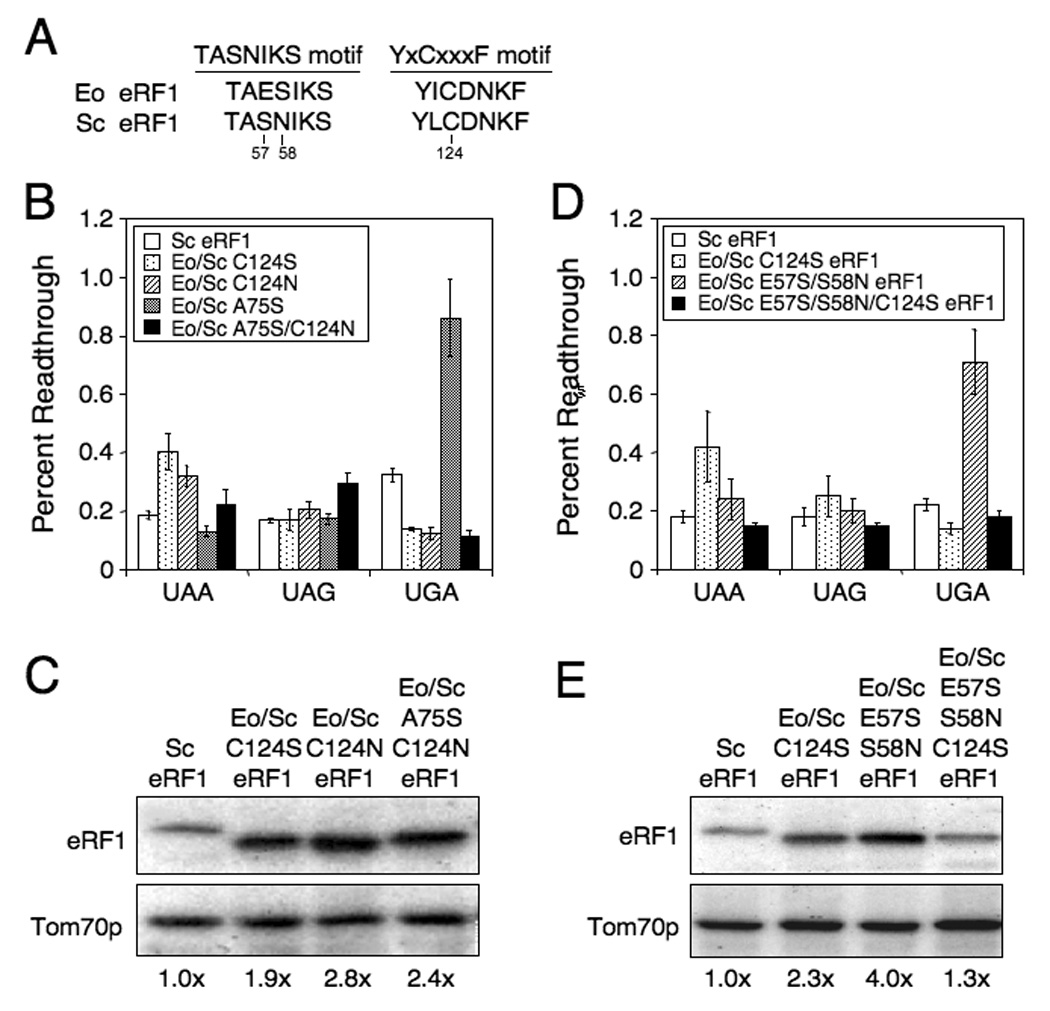
Efficiency of stop codon recognition mediated by Eo/Sc eRF1 suppressor mutants. (A) Alignment of TASNIKS and YxCxxxF motifs from Saccharomyces eRF1 and Euplotes eRF1. Numbering from Saccharomyces eRF1 is used. (B) Readthrough of stop codons measured in strains expressing wild type Sc eRF1 or Eo/Sc C124S, C124N, A75S or A75S/C124N eRF1. (C) Western blot quantitation of eRF1 levels from strains in (B). (D) Readthrough of stop codons measured in strains expressing wild type Sc eRF1 or Eo/Sc C124S, E57S/S58N, or E57S/S58N/C124S eRF1. (E) Western blot quantitation of eRF1 levels from strains in (D). All strains were grown in SM glucose medium at 35°C. Readthrough values are represented as mean ± SD.
