Abstract
In mesophilic prokaryotes, the DNA-binding protein HU participates in nucleoid organization as well as in regulation of DNA-dependent processes. Little is known about nucleoid organization in thermophilic eubacteria. We show here that HU from the hyperthermophilic eubacterium Thermotoga maritima HU bends DNA and constrains negative DNA supercoils in the presence of topoisomerase I. However, while binding to a single site occludes ∼35 bp, association of T. maritima HU with DNA of sufficient length to accommodate multiple protomers results in an apparent shorter occluded site size. Such complexes consist of ordered arrays of protomers, as revealed by the periodicity of DNase I cleavage. Association of TmHU with plasmid DNA yields a complex that is remarkably resistant to DNase I-mediated degradation. TmHU is the only member of this protein family capable of occluding a 35 bp nonspecific site in duplex DNA; we propose that this property allows TmHU to form exceedingly stable associations in which DNA flanking the kinks is sandwiched between adjacent proteins. We suggest that T. maritima HU serves an architectural function when associating with a single 35 bp site, but generates a very stable and compact aggregate at higher protein concentrations that organizes and protects the genomic DNA.
INTRODUCTION
Survival at extreme temperatures places unique demands on mechanisms that preserve integrity of the genomic DNA. Not only would duplex DNA more readily denature, but chemical processes that result in DNA damage, such as spontaneous depurination and strand breaks at apurinic sites, may be accelerated at elevated temperatures. A combination of evolutionarily conserved features therefore exist to prevent or combat the consequences of such stressors, including an increased G+ C-content of the genomic DNA, the existence of reverse gyrases that generate stabilizing positive DNA supercoiling, and the dynamic association with nucleoid-associated proteins.
In the mesophilic Escherichia coli, about a dozen distinct proteins associate with the nucleoid (1–3). These proteins include the essentially ubiquitously conserved HU and the structurally related integration host factor (IHF), sometimes referred-to as type II DNA-binding proteins, as well as H-NS, Fis and Dps (DNA protection during starvation), all of which are abundant and may reach micromolar concentration, depending on cell growth. Notably, most bacterial species do not encode homologs of all of these proteins. For instance, the hyperthermophilic eubacterium Thermotoga maritima, which has been phylogenetically placed as one of the deepest branching eubacterial lineages (4,5), does not encode homologs of IHF, Dps or H-NS. Unlike H-NS, whose role in DNA compaction is well documented, the extent to which E. coli HU participates in DNA compaction, particularly at higher concentrations, is a subject of ongoing debate (2,3,6–12).
While E. coli HU binds duplex DNA with low affinity and exhibits orders-of-magnitude higher affinity for flexible or prebent DNA, such as DNA with nicks or gaps or four-way junction structures, T. maritima HU (TmHU) binds both perfect duplex DNA and distorted DNA with nanomolar affinity (13–18). An additional feature that distinguishes TmHU is its ability to engage ∼37 bp of duplex, as opposed to the 9 bp site size for E. coli HU (18,19). TmHU and E. coli HU share 46% sequence identity and the partial structure of TmHU confirms its close structural relationship to mesophilic homologs (20–25). HU proteins function as dimers and bend DNA by partial intercalation of conserved prolines at two sites separated by 9 bp of duplex. The molecular basis for the distinct substrate specificities of these closely related proteins may reside in their differential ability to stabilize the proline-mediated DNA kinks, as evidenced also by the very different DNA bend angles imposed by Anabaena HU and IHF (22,23,26,27). Further, while IHF induces an only modest 10–15° dihedral angle and little net DNA unwinding, consistent with its inability to constrain negative DNA supercoiling, Anabaena HU induces both significant DNA underwinding and a variable 40–73° dihedral angle consistent with the negative supercoiling reported for several HU proteins (6,22,23,28–32). The type II DNA-binding protein encoded by Borrelia burgdorferi also constrains negative supercoils, but its DNA substrate lies largely in a single plane and the ability to constrain supercoils is likely a consequence of DNA underwinding near the kinks (25,32).
Little is known about the nucleoid structure in hyperthermophiles such as T. maritima. As in mesophiles, DNA compaction is expected to result from the combined action of topoisomerases and nucleoid-associated proteins; in addition to gyrase and topoisomerase I, T. maritima encodes a reverse gyrase—although the extent to which positive DNA supercoiling is generated is unclear—but as noted earlier, no homologs of the otherwise abundant proteins IHF, Dps and H-NS (6,33,34). The high-affinity binding of TmHU to perfect duplex DNA and the limited preference for alternate DNA substrates may presage a primary functional role in nucleoid organization and in maintaining genomic DNA integrity. We report here that TmHU exhibits different modes of interaction with short duplexes and with DNA capable of accommodating more than one TmHU protomer; the latter generates a complex in which the DNA is negatively supercoiled and effectively inaccessible, suggesting a role for TmHU in maintaining genomic DNA integrity.
MATERIALS AND METHODS
Electrophoretic mobility shift assay (EMSA) and quantitation of protein–DNA complexes
TmHU and Bacillus subtilis HU (HBsu) were overexpressed, purified and characterized as described (18,26). Oligomeric assemblies were assessed using glutaraldehyde-mediated crosslinking (27). Oligonucleotides used for preparation of 37- and 89-bp duplex DNA were purchased and purified by denaturing polyacrylamide gel electrophoresis. The top strand was 32P-labeled at the 5′-end with T4 polynucleotide kinase. Equimolar amounts of complementary oligonucleotides were mixed, heated to 90°C and slowly cooled to 4°C to form duplex DNA. The 242-bp duplex was obtained by PCR amplification of pU6L-BoxB (35) (primer sequences available on request), followed by 32P-labeling. Linear 462-bp DNA, used as competitor was prepared as a BglII digest. To generate 462-bp circles with negative or positive superturns, linearized DNA was incubated with T4 DNA ligase in the presence of either ethidium bromide (EtBr) or netropsin, followed by extensive phenol extraction and ethanol precipitation. Agarose gel electrophoresis of ligation products in the presence of 3.0 μg/ml chloroquine confirmed quantitative cyclization (i.e. no linear oligomers present) and the introduction of negative and positive superhelicity, respectively; the presence of chloroquine allows distinguishing negatively or positively supercoiled topoisomers that would otherwise migrate similarly.
EMSAs were performed using 10% (w/v) polyacrylamide gels (39:1 acrylamide:bisacrylamide) in TBE [45 mM Tris–borate (pH 8.3), 1 mM EDTA]. Gels were prerun for 30 min at 20 mA at room temperature before loading the samples with the power on. DNA and protein were mixed in binding buffer (20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.05% Brij58, 10 μg/ml BSA, 5% glycerol; except for experiments designed to assess the effect of salt concentration, in which the indicated concentration of KCl was used in place of NaCl), and each sample contained 1 fmol DNA in a total reaction volume of 10 μl for determination of equilibrium binding constants and 250 fmol for stoichiometric titrations. To assess inhibition of complex formation with linear or supercoiled DNA, complex formation with 37 bp DNA was performed as described earlier, followed by incubation with competitor for 10 min. Low-adherence tubes were used to minimize loss of protein sticking to surfaces. After electrophoresis, gels were dried and protein–DNA complexes were visualized and quantified by phosphoimaging, using software supplied by the manufacturer (ImageQuant 1.1). The region on the gel between complex and free DNA was considered as complex. DNA titrated with protein was fitted to f = fmax × [TmHU]n/(Kd + [TmHU]n) where [TmHU] is the protein concentration, f is fractional saturation, Kd is the apparent equilibrium dissociation constant, assuming protein binding to a single site and n is the Hill coefficient; for single-site binding, the Hill coefficient was set to 1. To determine the stoichiometry of binding, tangents were generated from data points in the upward slope and in the plateau and the stoichiometry of TmHU:DNA complex formation was extrapolated algebraically. For binding reactions including inhibitor, data were fit to a single exponential equation: normalized fractional complex formation = e−kL, where k is the exponential decay constant and L is the inhibitor concentration. All experiments were carried out at least in duplicate. Kd values are reported as the average ± the SEM.
EMSAs using agarose gels
One hundred nanograms of supercoiled, EcoRI-linearized or nicked pUC18, prepared by incubation of pUC18 with N.BstNBI (New England Biolabs Ipswich, MA, USA), was mixed with HU in 10 μl binding buffer [20 mM Tris–HCl, pH 8.0, 50 mM NaCl (unless specified otherwise), 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.05% Brij58, 10 μg/ml BSA, 5% glycerol). The entire reaction was loaded on a 0.5% agarose gel in TAE [40 mM Tris–acetate (pH 8.0), 1 mM EDTA]. N.BstNBI produces a single nick. DNA was visualized by EtBr staining following electrophoresis.
Supercoiling assays
Negatively supercoiled pGEM5 was relaxed by incubation at 37°C with Vaccinia Topoisomerase I in 50 mM Tris pH 8.0, 50 mM NaCl, 2.5 mM MgCl2 and 0.1 mM EDTA. Increasing concentration of TmHU was added and the reaction was allowed to continue for 60 min at 37°C. Reactions were terminated by addition of proteinase K to a final concentration of 0.17 mg/ml and incubation at 37°C for 1 h. DNA topoisomers were resolved by electrophoresis on 1% agarose gels in TBE at ∼3 V/cm for 17 h and visualized by EtBr staining. To distinguish positive and negative supercoiling, gels were soaked in chloroquine (3 μg/ml) after electrophoresis in the first dimension, turned 90° and the DNA resolved by electrophoresis in the second dimension. Chloroquine-soaked gels were stained with EtBr following electrophoresis in the second dimension. All experiments were performed at least in triplicate.
For ligase-mediated assays, nicked DNA was prepared by digestion of pUC18 with N.BstNBI. Reactions were performed in 1 × ligase buffer (T4 DNA ligase, New England Biolabs or Ampligase, Epicentre). TmHU was added to the DNA 45 min prior to the ligase. Reactions were performed and analyzed as described earlier, except that reactions with Ampligase were incubated at 65°C.
Cyclization assays
To assess DNA bending, plasmid pET5a was digested with BspHI to yield a 105 bp fragment, which was 32P-labeled at the 5′-end with T4 polynucleotide kinase. Fifty-three base pairs of DNA were obtained by AatII digestion of pcDNA3 (Invitrogen, Carlsbad, CA, USA). Reactions were initiated by addition of 80 U of T4 DNA ligase to a final volume of 10 μl. Reactions containing 100 fmol DNA and 50 fmol TmHU were incubated in 1 × binding buffer [20 mM Tris–HCl pH 8.0, 10 mM MgCl2, 50 mM NaCl, 0.1 mM Na2EDTA, 0.1 mM DTT, 0.05% (w/v) Brij58, 100 μg/ml BSA] with 0.5 mM ATP at room temperature for 1–30 min and terminated with 5 μl of 75 mM EDTA, 3 mg/ml proteinase K, 15% glycerol, and bromophenol blue and xylene cyanol, followed by a 15 min incubation at 55°C. Cyclized DNA was identified by its resistance to digestion by Exonuclease III. Reactions were analyzed on 8% polyacrylamide gels [39:1 (w/w) acrylamide: bisacrylamide] in TBE. After electrophoresis, gels were dried and ligation products visualized and quantified by phosphorimaging.
Two-dimensional DNA footprinting
DNA probes were prepared by 5′-end-labeling one strand prior to annealing the complementary strands of the 89 bp looped DNA. Protein–DNA complexes were formed in a 10 μl volume for 5 min at room temperature under reaction conditions as described earlier. Each reaction contained 500 fmol of DNA. DNase I (adjusted to yield <10% DNA cleavage to ensure single hit) was added to each reaction which was terminated after 30 s at room temperature by loading samples onto a running nondenaturing 8% polyacrylamide gel in TBE. Protein–DNA complexes were isolated and the DNA was passively eluted from gel slices, extracted with phenol–chloroform, precipitated with ethanol and resolved on 15% polyacrylamide sequencing gels. Sequence positions were identified by comparison to an A + G ladder. Density profiles were obtained by phosphorimaging.
DNA protection assays
One hundred nanograms of pUC18 DNA was incubated with 0.1–0.5 µM TmHU or HBsu in a 10 µl total reaction volume at room temperature for 20 min in the presence of 1 × binding buffer and 1 × DNase I buffer (New England Biolabs) followed by incubation with 0.1 U DNase I for 1, 3 or 5 min. Reactions were terminated by adding 5 µl DNase I stop buffer (10% SDS, 20 µM EDTA) and incubating for 10 min at room temperature. In control reactions, no TmHU or HBsu was added. To assess the effect of mechanical stress on the DNA, DNase I stop buffer was added to the reaction by pipetting once, by pipetting 20 times or by vortexing for 30 s. Reactions were loaded onto 1% agarose gels and viewed by EtBr staining following electrophoresis. DNA was quantitated by densitometry using an Alphaimager (Alpha Innotech Corporation, San Leandro, CA, USA).
RESULTS
TmHU bends DNA
Based on sequence and structural conservation, TmHU is expected to exist as a dimer, an expectation borne out by examination of cross-linked species in solution (see Supplementary Figure S1, which also documents the >95% purity of the protein preparation, as judged by Coomassie-staining of SDS–PAGE gels). No monomeric species are detectable regardless of protein concentration, but defined higher-order oligomers are seen at higher protein concentrations, with assemblies corresponding to two, three or four copies of the TmHU dimer distinguishable.
A conserved feature of type II DNA-binding proteins is their ability to bend DNA by partial intercalation of two proline residues at two positions separated by 9 bp of duplex (22,23). This DNA-intercalating proline is conserved in TmHU (for sequence alignment, see Supplementary Figure S2). However, despite significant levels of sequence and structural conservation, the ability to bend and/or supercoil duplex DNA is not uniformly conserved among type II DNA-binding proteins and cannot be assumed based solely on sequence conservation (22,23,27–33,36). Using DNA duplexes of varying length, TmHU was previously shown to interact optimally with 37 bp perfect duplex DNA and to exhibit lower affinity binding to shorter duplexes (18). Accordingly, TmHU should introduce a pair of proline-mediated DNA kinks in 37-bp DNA at a spacing of 9 bp, with the concomitant wrapping of flanking DNA about the protein surface, akin to the interaction between IHF and its DNA target (18,22). Such interaction predicts bending of duplex DNA. Ligase-mediated cyclization assays measure the efficiency with which T4 DNA ligase mediates ring closure of DNA fragments that are shorter than the persistence length. While 105-bp DNA fails to yield intramolecular ligation products (Figure 1), incubation with TmHU leads to the rapid formation of circular DNA products, as evidenced by their resistance to digestion with Exonuclease III, documenting the ability of TmHU to bend the DNA duplex and promote intramolecular ring closure. In either case, cyclization of linear dimers occurs with longer time of incubation. Fifty-three base-pair duplex was not cyclized in the presence of TmHU, instead cyclization of 106 bp linear dimers was enhanced (data not shown).
Figure 1.
Thermotoga maritima HU promotes DNA bending. (A and B) DNA cyclization assay in the absence or presence of TmHU as indicated above the panels. Lane 1 shows 105 bp linear duplex DNA. Reactions in lanes 2–8 contained 105-bp DNA incubated with T4 DNA ligase, and reactions in lanes 10–16 additionally contained 50 fmol TmHU. Reactions in lanes 2 and 10 were incubated with T4 DNA ligase for 1 min, reactions in lanes 3 and 11 for 2 min, reactions in lanes 4 and 12 for 8 min, reactions in lanes 5 and 13 for 11 min, reactions in lanes 6 and 14 for 14 min, reactions in lanes 7 and 15 for 17 min and reactions in lanes 8 and 16 for 30 min. Reactions in lanes 9 and 17 were treated with Exonuclease III following incubation with ligase for 30 min. The original 105 bp DNA, dimeric ligation product, intramolecular cyclization product and linear oligomers are identified at the right.
TmHU constrains negative DNA supercoils in the presence of topoisomerase I
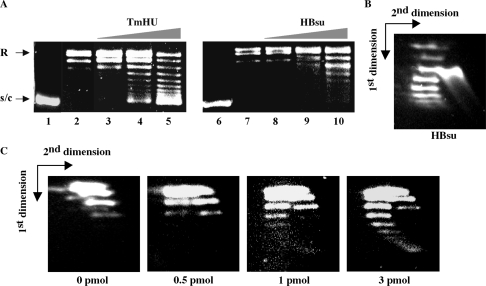
DNA supercoiling was assessed using covalently closed, relaxed plasmid DNA, prepared by incubation of supercoiled pGEM5 with Vaccinia topoisomerase I (Figure 2). The efficiency of DNA supercoiling was compared to that of HBsu, the HU homolog from the mesophilic B. subtilis, previously shown to produce negative DNA supercoiling (27,29,30). TmHU and HBsu exhibit 50% sequence identity. TmHU supercoils relaxed plasmid DNA more efficiently than HBsu; at any given protein:DNA ratio, ▵Lk is greater for TmHU than for HBsu, consistent with higher affinity binding (18,27). Plasmid incubated with TmHU exhibits a bimodal distribution of topoisomers and ▵Lk is not linearly dependent on [TmHU].
Figure 2.
TmHU negatively supercoils DNA in the presence of topoisomerase I. (A) Supercoiling by TmHU (left panel) and HBsu (right panel). Lanes 1 and 6 contain supercoiled DNA and lanes 2 and 7 pGEM5 relaxed by incubation with Vaccinia topoisomerase I. Lanes 3–5 and 8–10 represent reactions in which 0.5, 1.0 and 3.0 pmol of TmHU or HBsu, respectively, were added to relaxed DNA, followed by a further 60 min incubation. (B and C) Two-dimensional agarose gel electrophoresis. Gels were soaked in chloroquine after electrophoresis in the first dimension, turned 90°, and positively and negatively supercoiled DNA topoisomers were separated by electrophoresis in the second dimension. (B) Negative supercoiling constrained by 10 pmol HBsu. The species migrating ahead of the supercoiled topoisomers is linear DNA. (C) Increasing negative superhelicity is seen with increasing concentrations of TmHU. Protein concentrations are indicated below each panel. Already positively supercoiled DNA acquires additional positive superhelical turns and moves faster after staining with chloroquine, forming the right branch of the arc, and negatively supercoiled DNA looses superhelicity and moves slower. The directions of the first and second dimensions are indicated by arrows.
The direction of DNA supercoiling was determined by 2D agarose gel electrophoresis. When supercoiled DNA is relaxed by Vaccinia topoisomerase I in the presence of Mg2+, a thermal distribution of DNA topoisomers is generated that is characterized by modest positive superhelicity due to the effect of Mg2+ on the free energy of DNA supercoiling (Figure 2C, left panel). As previously reported, HBsu constrains negative supercoils (Figure 2B). On addition of increasing concentrations of TmHU, an increase in negative superhelicity is also seen; however, the positively supercoiled topoisomers remain (Figure 2C, right panels). The retention of DNA topoisomers with positive superhelicity suggests that TmHU, when added to the mixture of relaxed or positively supercoiled DNA generated by the topoisomerase, may bind preferentially to the positively supercoiled DNA compared to relaxed DNA, preventing its relaxation by topoisomerase I and leading to the formation of two coexisting populations of topoisomers. In absence of Mg2+, only negatively supercoiled topoisomers are seen (data not shown).
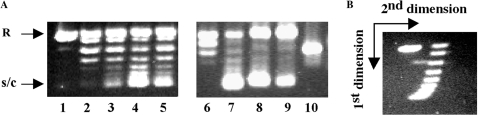
The affinity of TmHU for a single 37-bp site is constant within a temperature range of 4–45°C (18). To assess whether DNA compaction by TmHU is likewise insensitive to temperature, nicked DNA was ligated in the presence of TmHU with T4 DNA ligase at room temperature or with ampligase at 65°C, closer to the physiological growth temperature of T. maritima. While T4 DNA ligase and ampligase religate the nicked DNA with comparable efficiency (compare Figure 3A, lanes 2 and 6), lower concentrations of TmHU are sufficient to constrain supercoiling in the presence of ampligase (compare lanes 3 and 7), suggesting that TmHU constrains supercoils more efficiently at physiological temperatures. In either case, higher concentrations of TmHU are inhibitory, suggesting that DNA becomes inaccessible to the ligase. At room temperature, 5 pmol TmHU efficiently complexes all DNA (lane 10) and the faster migration of the complex compared to free-nicked DNA indicates significant DNA compaction. That only negatively supercoiled topoisomers are seen (Figure 3B) also indicates that the retention of positively supercoiled topoisomers in topoisomerase I-mediated supercoiling assays is not due to effects of Mg2+ on protein binding.
Figure 3.
Temperature-dependent supercoiling. (A) Ligation of nicked pUC18 by T4 DNA ligase (room temperature; lanes 2–5) or ampligase (65°C; lanes 6–9). Lane 1, DNA only. Lanes 2 and 6, no TmHU. Lanes 3–5 and 7–9, reactions in which 2.5, 5.0 and 10.0 pmol TmHU were added to nicked pUC18 prior to addition of T4 DNA ligase (lanes 3–5) or ampligase (lanes 7–9). Lane 10, nicked DNA incubated with 5 pmol TmHU. (B) Two-dimensional agarose gel electrophoresis of nicked pUC18 ligated by T4 DNA ligase in the presence of 2.5 pmol TmHU. The gel was soaked in chloroquine after electrophoresis in the first dimension, turned 90° and DNA topoisomers were separated by electrophoresis in the second dimension. The directions of the first and second dimensions are indicated by arrows. That the tails of supercoiled topoisomers bend in the opposite direction compared to that seen in the figure reflects a more efficient introduction of positive superhelicity in pGEM5 by chloroquine.
TmHU binds preferentially to supercoiled DNA
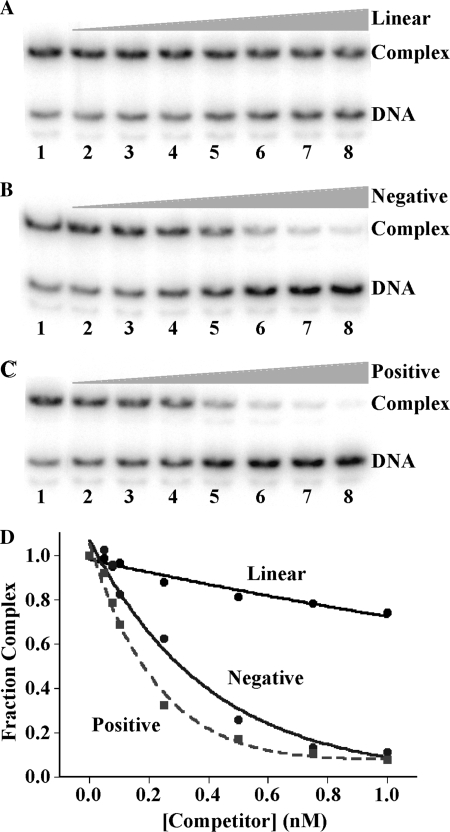
To address explicitly the inference that TmHU binds preferentially to supercoiled DNA, DNA with positive or negative superhelicity was prepared using a 462 bp BglII fragment, circularized with T4 DNA ligase in the presence of either EtBr or netropsin. These compounds introduce underwinding and overwinding of the DNA, respectively, that is, they alter the twist of the DNA. Therefore, extraction of the compounds following ligation (conditions under which ▵Lk remains unchanged) results in compensatory negative or positive writhe, respectively, as the twist is restored to an average ∼10.5 bp/turn (37,38). As shown in Figure 4, TmHU complex formation with 37-bp DNA is only modestly affected by challenge with unlabeled 462 bp linear DNA (top panel), while supercoiled DNA competes more efficiently for binding, consistent with preferred binding to supercoiled DNA of either handedness.
Figure 4.
TmHU binds preferentially to supercoiled DNA. (A–C) EMSA in which 37-bp DNA was incubated with TmHU prior to addition of 462 bp competitor DNA. Lanes 1, no competitor. Lanes 2–8, increasing concentration of competitor [identical concentrations of each competitor with concentrations indicated in (D)]. (A) Competition with linear 462 bp DNA, (B) negatively supercoiled 462 bp DNA and (C) positively supercoiled 462-bp DNA. DNA and complex is identified at the right. Relative fraction of complex as a function of competitor concentration is shown in (D).
The wrapping of DNA about the surface of other HU homologs has been shown to involve disruption of surface salt bridges, and the wrapping path proposed to be defined by a specific pattern of surface salt bridges (39,40). Such coupled disruption of surface salt bridges is experimentally evidenced by a very modest salt-dependence of binding. EMSAs using 37 bp duplex DNA, which can accommodate only one TmHU dimer [Figure 4; (18)], shows that the apparent dissociation constant Kd is only very modestly dependent on salt concentration, with Kd = 1.0 ± 0.1 nM and 1.5 ± 0.4 nM in 50 mM and 250 mM KCl, respectively (data not shown). This limited salt dependence of binding is consistent with a coupled disruption of surface salt bridges upon TmHU–DNA complex formation.
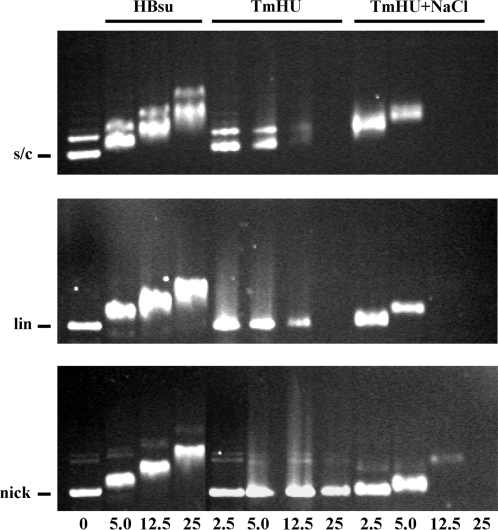
While binding to a single site appears not to be significantly affected by salt, association of TmHU with plasmid DNA does reveal such dependence. At 50 mM NaCl, titration of supercoiled DNA with TmHU results in an only modest retardation of the resulting complex, but a gradual reduction in the intensity of EtBr staining, suggesting that the DNA becomes inaccessible to EtBr (Figure 5, top row, middle panel). Addition of SDS to disrupt TmHU–DNA complex formation prior to electrophoresis quantitatively restores free DNA (not shown). While 12.5 pmol TmHU renders supercoiled DNA nearly inaccessible to EtBr, higher concentrations are required to saturate linear or nicked DNA, consistent with preferred binding to supercoiled DNA. Since complexes become progressively less accessible to EtBr, we surmise that the almost unaltered mobility reflects DNA compaction counteracting retardation of the complex in the gel (note that electrophoresis under different conditions results in complexes migrating faster than the free DNA [Figure 3A, lane 10)]. Notably, raising the [NaCl] to 200 mM causes well-defined TmHU–DNA complexes to migrate slower and to become inaccessible to EtBr at lower TmHU concentrations (rightmost panels). In contrast, association of DNA of either topological form with HBsu is qualitatively similar, complexes migrate significantly slower than free DNA and the DNA remains accessible to EtBr (leftmost panels).
Figure 5.
DNA compaction by TmHU is salt-dependent. Agarose gel electrophoresis of HBsu, TmHU at 50 mM NaCl (TmHU) or TmHU at 200 mM NaCl (TmHU + NaCl) binding to supercoiled (s/c; top row), linear (lin; middle row), or nicked (nick; bottom row) pUC18 DNA. Protein concentrations, identical for all rows, are indicated at the bottom. Reactions in lanes 1 contain no protein, with the position of the free DNA identified at the left. DNA is stained with EtBr after electrophoresis; DNA in reactions with higher concentrations of TmHU fails to stain.
TmHU exhibits a distinct mode of interaction with DNA capable of accommodating multiple protomers
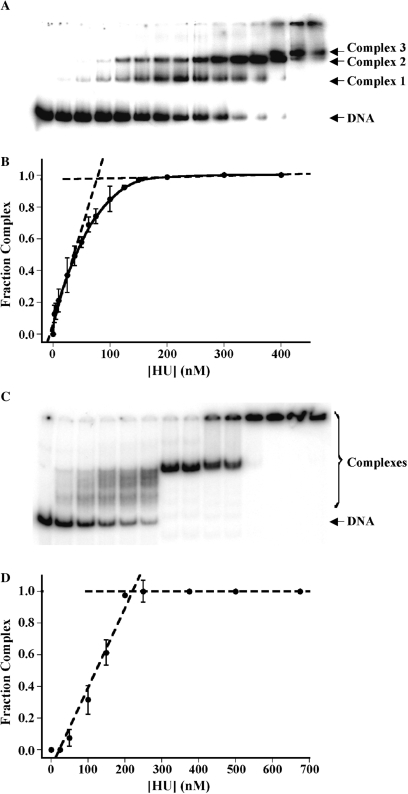
Since interaction of TmHU with 37-bp DNA yielded only a single complex, a stoichiometry of 1:1 was inferred for 37-bp DNA (18). A longer site size was also indicated compared to E. coli HU, which was previously seen to form three complexes with 29-bp DNA, consistent with a 9-bp site (19). However, the previously reported analysis did not rule out the possibility that TmHU might associate with DNA in an oligomeric form other than the expected dimer. We therefore performed stoichiometric titrations ([DNA] >Kd) of TmHU with DNA of varying length to determine the occluded site size. Interaction of TmHU with 37 bp DNA yields a single complex (Figure 4). When the fraction of complex formation was plotted against [TmHU] and tangents were drawn in the upward slope and the saturation plateau, a break point was observed reflecting saturation of the DNA at a protein:DNA stoichiometry of 1:1, corresponding to an occluded site size of 35.5 bp/TmHU (data not shown). This result is also consistent with the previously reported decrease in binding affinity for TmHU interacting with DNA shorter than 37 bp (18).
The determination of DNA site size for TmHU in DNA longer than 37 bp suggests an apparent reduction in the occluded site size with increasing duplex length. As shown in Figure 6A, interaction with 89-bp DNA yields three complexes of defined mobility. However, the number of complexes observed cannot be necessarily correlated to the number of protein molecules bound, for the following reason. TmHU bends DNA (Figure 1); for the short, 37-bp DNA, electrophoretic mobility is primarily determined by the molecular mass of the protein–DNA complex and not by the shape of the DNA. However, as the DNA gets longer, a different electrophoretic mobility is seen for otherwise stoichiometrically equivalent complexes in which a DNA-bending protein is bound at the end or at the center of the DNA molecule, with end-bound DNA migrating fastest. The distinct complexes seen on incubation of 89-bp DNA with TmHU may therefore reflect both differences in mobility due to the position of the protein relative to the DNA ends as well as the stoichiometry. When the fraction of complex formation with 89-bp DNA was plotted against [TmHU], a break point was observed reflecting saturation of the DNA at a protein concentration corresponding to 28.8 bp/TmHU (Figure 6B).
Figure 6.
TmHU exhibits an apparent decrease in occluded site size with increasing duplex length. (A) EMSA under stoichiometric conditions with 89-bp DNA and (B) the corresponding binding isotherm, in which the intercept of tangents to the saturation plateau and the initial slope reflects saturation of the DNA. (C) EMSA under stoichiometric conditions with 242-bp DNA and (D) binding isotherm reflecting saturation of 242-bp DNA by TmHU; note that all DNA still available to bind TmHU, not just unbound DNA, was considered as ‘free DNA’. For (A) and (C), complexes and free DNA are identified at the right. Protein concentrations are reflected in the corresponding binding isotherms.
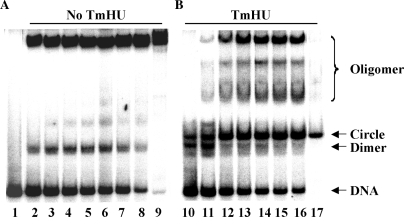
With 242-bp DNA, substoichiometric concentrations of TmHU yield complexes of varying mobility, resulting in what appears as a smear in the gel. The mode of TmHU–DNA interaction, that is, its nonsequence-specific binding and its introduction of a significant DNA bend, combined with the observation noted above about the differential migration of protein–DNA complexes in which a bend is introduced at different positions relative to the DNA ends, allows some deductions about the likely nature of these complexes. TmHU does not bind DNA sequence specifically, implying the existence of numerous overlapping sites in the DNA. As protein bound at the center will result in a complex with much reduced mobility compared to protein bound at the DNA ends, with protein bound at any position in-between yielding a complex of intermediate mobility, a population of complexes will result that all have different mobility, resulting in the appearance of a smear in the gel (Figure 6C).
At a protein concentration corresponding to approximately two TmHU dimers per DNA, a single complex of well-defined mobility is formed. We infer that it may be the ability of TmHU to self-associate that promotes the formation of a complex in which two TmHU protomers associate such that the two bends oppose each other, resulting in little or no net DNA bend and equivalent electrophoretic mobility regardless of the position of the bound proteins relative to the DNA ends. An alternate mechanism that may promote such cooperative interaction between two DNA-bending proteins depends not on physical interaction, but instead on cooperative binding mediated by deformation of the intervening DNA (41). Regardless of which mechanism underlies the observed complex formation, the association of two TmHU protomers evidently results in a complex with defined electrophoretic mobility. A further increase in [TmHU] results in further retardation of the complex and its retention in the well. At saturation, a stoichiometry of ∼9:1 is seen, with each TmHU dimer occluding 26.8 bp (Figure 6D). These data suggest a closer packing of TmHU on DNA of sufficient length to accommodate multiple protomers.
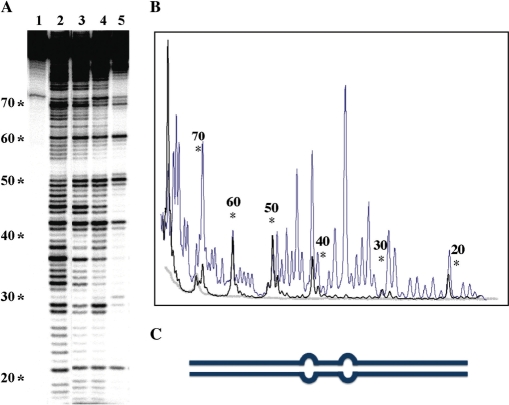
To assess the basis for the apparent reduction in occluded site size on incubation of TmHU with DNA capable of accommodating more than a single TmHU dimer, DNase I footprinting was performed. TmHU has a modestly (∼4-fold) higher affinity for DNA with a set of 4-nt loops separated by 9 bp of duplex compared to perfect duplex DNA (18). This enhanced affinity was expoited to position TmHU preferentially on 89-bp DNA containing tandem loops at the center of the construct. As for 89 bp perfect duplex DNA (which is seen in Figure 6A), three distinct complexes are seen with the 89 bp looped DNA (not shown). Two-dimensional DNase I footprinting was selected because TmHU has only modest preference for the loops present in this DNA construct; in solution, a mixed population of complexes would therefore exist. Complexes sufficiently distinct to elicit differential electrophoretic mobility may be separated by EMSA, and DNA isolated from each complex may be subjected to denaturing gel electrophoresis to identify cleavage products. This experiment shows that the fastest migrating complexes (corresponding to Complex 1 and 2 in Figure 6) must be ensemble averages of TmHU bound at various locations, as the protection patterns are quite dispersed and incomplete (Figure 7A; this observation is also consistent with the above noted caveat, that the relative migration of complexes with longer DNA likely reflects both the position of the protein relative to the DNA end as well as stoichiometry). In contrast, the slowest migrating complex (Complex 3) reveals complete protection interrupted by localized cleavage centered at positions 21, 30, 42, 50, 60 and 69 (Figure 7B). Two sites (42 and 50) correspond to the positions of the loops, suggesting preferential binding of one TmHU dimer at this position. The periodicity of the cleavage sites suggests ordered placement of TmHU.
Figure 7.
TmHU condenses DNA. (A) Two-dimensional DNase I footprinting with 89 bp looped DNA. The position of loops corresponds to bases 40–41 and 49–50 (positions are identified to the left of the sequencing gel). Reactions in (A) were: lane 1, DNA only; lane 2, DNA treated with DNase I in absence of TmHU; lanes 3–5 TmHU:DNA complexes 1, 2 and 3, respectively, as identified in (A). All reactions were subjected to EMSA and either free DNA (lanes 1 and 2) or TmHU–DNA complex (lanes 3–5) isolated from the gel as described in Materials and methods section prior to denaturing gel electrophoresis to separate cleavage products. (B) Densitometry profile of uncut DNA (gray line), DNase I-digested DNA (blue) and TmHU:DNA complex 3 (black). (C) Cartoon illustration of the 89 bp looped DNA.
TmHU protects DNA against degradation
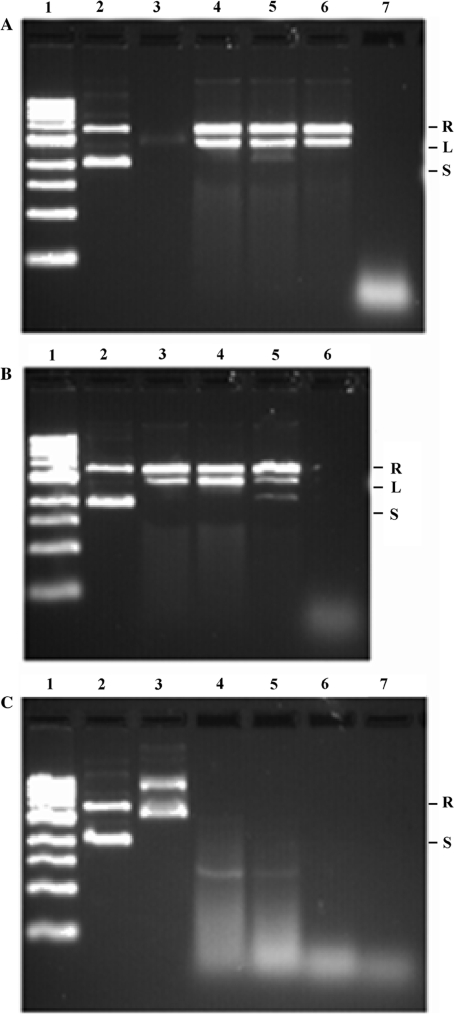
As the footprinting pattern suggested DNA protection at high protein density, we examined this property further. When plasmid was incubated with TmHU, protection against DNase I-mediated degradation was evident (Figure 8A, lanes 4–6). Using an initial mixture of ∼70% supercoiled and ∼30% relaxed DNA, incubation with DNase I in the presence of TmHU resulted in ∼55% relaxed and ∼45% linear species. Remarkably, recovery of total DNA was essentially 100% even after 5 min incubation, indicating a very stable complex in which no more than one double strand break and possibly multiple nicks are present. To ascertain whether the plasmid DNA in the complex is primarily nicked by DNase I and the double strand breaks are due to handling of the DNA after recovery from the complex, the DNA was subjected to increasing levels of mechanical stress, from pipetting once to vortexing extensively (Figure 8B, lanes 3–5). Additional double strand breaks were introduced by mechanical stress; however, the essentially quantitative recovery suggests a limited number of nicks. Under similar conditions, HBsu does not protect the DNA (Figure 8C).
Figure 8.
TmHU protects DNA from degradation. (A) Protection of plasmid pUC18 by TmHU. Lane 1, DNA marker; lane 2, pUC18 only; lane 3, TmHU–pUC18 complex; note that DNA fails to stain with EtBr due to occlusion by TmHU; lanes 4–6, TmHU–DNA complexes treated with 0.1 U DNase I for 1 min (lane 4), 3 min (lane 5) and 5 min (lane 6); lane 7, pUC18 incubated with DNase I for 3 min. Reactions in lanes 4–7 were terminated with SDS-containing stop-buffer to recover the DNA. (B) Recovered DNA was subjected to increasing levels of mechanical stress. Lane 1, DNA marker; lane 2, pUC18 only; lanes 3–5, DNA recovered after 3 min incubation with DNase I in the presence of TmHU mixed by pipetting once (lane 3), pipetting 20 times (lane 4) and vortexing for 30 s (lane 5); lane 6, DNA incubated with DNase I. (C) No DNA protection by HBsu. Lane 1, DNA marker; lane 2, pUC18 only; lane 3, HBsu–pUC18 complex; lanes 4–6, HBsu-pUC18 complexes treated with 0.1U DNase I for 1 min (lane 4), 3 min (lane 5) and 5 min (lane 6); lane 7, pUC18 treated with DNase I for 3 min. Reactions in lanes 4–7 were terminated with SDS-containing stop-buffer to recover the DNA. Relaxed (R), Linear (L) and supercoiled (S) DNA identified at the right.
While HBsu is unable to protect DNA in vitro, it has been previously shown to compact and organize genomic DNA in vivo when overexpressed in B. subtilis (42). While in vivo DNA compaction due to HU binding would make intuitive sense, noting that E. coli lacking HU exhibits more disperse nucleoids, a contribution from indirect effects on nucleoid compaction due to inhibition of RNA or protein synthesis cannot be ruled out (43). This caveat notwithstanding, a similar compaction is seen on overexpression of either HBsu or TmHU in E. coli, as evidenced by DAPI staining of the nucleoids (see Supplementary Figure S3). Thus, while both proteins are capable of contributing to DNA compaction in vivo, only TmHU generates complexes that resist nuclease digestion (Figure 8).
DISCUSSION
Unique demands on a hyperthermophilic HU homolog
Little is known about the nucleoid structure in hyperthermophiles. Protection of genomic DNA against thermal denaturation in hyperthermophilic eubacteria or archaea sometimes involves an increased G + C-content; however, this route is not used by T. maritima whose genome has an average G + C-content of only 46% (44). Another characteristic of hyperthermophiles is that they encode a reverse gyrase capable of introducing stabilizing positive supercoiling in the DNA. Thermotoga maritima is no exception. However, plasmid isolated from Thermotoga sp. was reported to be negatively supercoiled, and topoisomerase I activity was reported to be the primary topoisomerase activity in T. maritima crude extract (34,45). That positive DNA supercoiling may contribute to DNA stability is also indicated by reports that some hyperthermophilic archaea encode histones with the capacity to constrain DNA into both positive and negative supercoils; archaeal histones are closely related to eukaryotic histones, but assemble into tetramers for DNA binding (46–49). The ability to constrain DNA into positive toroidal supercoils has been evolutionarily conserved in the eukaryotic histone (H3-H4)2 tetramer that may undergo a reorientation of the H3-H3 interface that leads to a switch of wrapped DNA from a left- to a right-handed superhelix (50). Proteins with the capacity to constrain positive DNA supercoils have likewise been reported in the crenarchaeota, which do not encode histone homologs (51).
In T. maritima, TmHU is predicted to be the major nucleoid-associated protein. We report here that TmHU exhibits three unique features, not characteristic of homologs encoded by mesophiles such as E. coli and B. subtilis: (i) binding to a 37 bp single site for which a large bend angle is predicted based on structures of other HU–DNA complexes; (ii) constraint of negative supercoiling in the presence of topoisomerase I and the stabilization of existing positive supercoiling, the latter consistent with a role in stabilizing DNA against thermal denaturation and (iii) protection of DNA by formation of TmHU–DNA filaments in which the DNA is effectively inaccessible, a feature consistent with a role in genomic DNA organization and protection. Thus, TmHU exhibits two features that may contribute uniquely to protection of genomic DNA in a hyperthermophilic environment, the ability to stabilize positive supercoils and the ability to render DNA effectively inaccessible, thereby protecting it from both thermal denaturation as well as temperature-dependent degradation. Notably, that TmHU is seen to constrain only negative DNA supercoils in the presence of topoisomerase I is consistent with the previously observed negative supercoiling of plasmid isolated from Thermotoga sp. (34), and it suggests that TmHU may stabilize structures that would otherwise be destabilizing at higher temperatures, such as the negative supercoiling that emerges in the wake of a transcribing RNA polymerase. What follows is a more explicit mechanistic consideration of each functional property.
TmHU may introduce a large DNA bend and serve an architectural function in its binding to a single site
Thirty-seven base pairs of DNA, which corresponds to the length of DNA duplex that may be wrapped about the surface of TmHU, can accommodate a single TmHU dimer; a single complex is observed and stoichiometric titrations specify occlusion by one TmHU protomer (18). At low protein concentrations, association with a 37-bp site in longer DNA is also likely [particularly considering that TmHU appears not to bind cooperatively (Figure 6C)]. Under these circumstances, TmHU must bend its target DNA to allow contacts between DNA distal to the kinks and the protein surface, presumably generating ∼160° bends as seen in the structures of E. coli IHF or B. burgdorferi Hbb with 35-bp duplexes (22,25). In contrast, the bend angle imposed by Anabaena HU, which engages a shorter DNA site, is variable (105–139°) (23). What is unique about TmHU is that it is the only member of the HU/IHF family that has been shown to engage a 37-bp site in duplex DNA without sequence specificity; for instance, association of E. coli IHF with nonspecific sites is accompanied by significant reductions in both affinity and occluded site size (39). As T. maritima encodes only one member of this protein family, it is possible that TmHU is required as an architectural factor in specific DNA-dependent processes, such as DNA replication. While TmHU appears not to exhibit sequence specificity, its preferred binding to supercoiled DNA may yet direct it to localized DNA sites, such as the apex of a plectonemic superhelix (Figure 4) (52).
TmHU constrains negative DNA supercoiling, yet stabilizes existing positive DNA supercoils
TmHU restrains negative DNA supercoils in the presence of topoisomerase I (Figure 2). DNA supercoiling can be realized either by altering the twist of the double helix (Tw) or by modifying the writhe (Wr) by out-of-plane bending of the helix axis. When enzyme-mediated rejoining of the DNA strands takes place, the relative contributions of Tw and Wr to the ΔLk at that very moment in time are fixed. For both E. coli IHF and B. burgdorferi Hbb, their 35-bp DNA substrates exhibit little out-of-plane bending, whereas the shorter duplex in complex with Anabaena HU features large, yet variable dihedral angles consistent with negative supercoiling (22,23,25). Indeed, it has been suggested that binding to a shorter DNA site is associated with larger dihedral angles between distal DNA segments (25). Negative DNA supercoiling would also result from DNA underwinding; for both Anabaena HU and B. burgdorferi Hbb, net DNA underwinding is seen for the respective duplexes, while IHF induces little DNA underwinding, consistent with its inability to constrain DNA supercoils. For TmHU, we speculate that supercoiling may result largely from a change in Tw; the small sample size notwithstanding, the ability to constrain DNA supercoils appears to correlate with DNA underwinding, not with out-of-plane bending. That TmHU binds preferentially to supercoiled DNA of either handedness (Figure 4) is also consistent with a wrapping path that does not strongly favor out-of-plane bending; in this circumstance, TmHU likely binds the apex of a plectonemic superhelix generated by intramolecular ring closure. In absence of bending, the thermal effect on DNA unwinding is –0.012°/bp/°C, resulting in 34.4°/bp at RT and 33.7°/bp at 80°C (53–55). Facilitated supercoiling at elevated temperatures would therefore be an anticipated outcome if TmHU unwinds DNA, consistent with the enhanced constraint of negative supercoils at 65°C (Figure 3).
TmHU binds preferentially to supercoiled DNA (Figures 2 and 4). If positively supercoiled DNA topoisomers are retained in the presence of topoisomerase I due to preferred binding by TmHU (Figure 2), then a change in topoisomer distribution might be expected as a function of protein concentration or time of incubation. Since TmHU constrains negative supercoils, compensatory positive supercoils would form in naked DNA, to which TmHU should also preferentially bind, predicting that both positive and negative topoisomers should increase with [TmHU]. This is not observed. However, the quantitative recovery of plasmid DNA following DNase I digestion suggests that TmHU does not dissociate measurably to permit access of DNase I to the DNA (Figure 8). We therefore surmise that reaction mixtures with TmHU and topoisomerase I are not at equilibrium, and that TmHU, once bound dissociates only slowly, thus stabilizing the existing distribution of topoisomers. Accordingly, TmHU may well stabilize positively supercoiled DNA in vivo if such species are generated, for example, by passage of DNA or RNA polymerases. Notably, an equivalent retention of positively supercoiled topoisomers is not observed in the presence of HBsu (Figure 2B). For a homodimeric E. coli HUα containing two amino acid substitutions, constraint of positive supercoils was reported (12). However, this phenomenon was observed only at high protein concentrations and was inferred to occur only upon generation of octameric assemblies of the mutant homodimer. Since the level of positive supercoils retained in the presence of TmHU does not change with increasing protein concentration (Figure 2C), we have no reason to suspect its dependence on the formation of higher order oligomeric assemblies of TmHU.
TmHU forms compact structures in which DNA is effectively inaccessible
It is likely not just thermal DNA denaturation, but thermally enhanced DNA degradation against which hyperthermophiles need protection. We propose that TmHU is contributing to such protection, a hypothesis that is based on the collective properties of DNA in complex with TmHU. Significant DNA compaction is suggested by the observation that TmHU–DNA complexes migrate faster than free plasmid DNA under certain electrophoretic conditions (Figures 3 and 5, lane 10). This is markedly different from the association with HBsu, which leads to complexes with significantly reduced electrophoretic mobility. It is also remarkable that DNA in complex with TmHU becomes inaccessible to EtBr, while complexes with HBsu remain fully stainable (Figure 5). That incubation of TmHU–DNA complexes with DNase I only results in modest DNA nicking (Figure 8) further suggests that the complex, once formed, dissociates only slowly. In contrast, DNA in complex with HBsu is extensively degraded. This may reflect reduced stability of the HBsu–DNA complex, although we note that complexes with E. coli HU were seen to remain stable in absence of challenge with competitor DNA (56) and that complexes with both TmHU and HBsu remain stable upon 10-fold dilution with buffer (data not shown). An alternate explanation is that HBsu, like Anabaena HU (23), introduces a range of bend angles, resulting in a ‘breathing’ complex in which DNase I may access the DNA. The latter interpretation is also consistent with the proposal by Rice and coworkers that binding to a shorter DNA site is associated with larger and variable dihedral angles between distal DNA segments, perhaps resulting in a more loosely packed complex; the occluded DNA site for HBsu was estimated at 10–13 bp (25,27).
What is different in the association of TmHU with long DNA that results in the DNA becoming effectively inaccessible, both to EtBr staining and to nucleolytic degradation? Stability of the complex per se may not be the main difference, considering that high concentrations of E. coli HU form stable complexes that dissociate only slowly when equilibrated against buffer solution; however, addition of competitor DNA resulted in immediate complex disassembly (56). It is possible that complexes with TmHU are more stable under conditions of excess ligand in solution, as suggested by the retention of positively supercoiled DNA topoisomers even in presence of excess relaxed DNA (Figure 2). Does the level of DNA compaction differ? That complexes with TmHU may migrate faster than free DNA speaks to such compaction (Figure 3, lane 10). In E. coli, DNA compaction by H-NS has been well documented (3,9). Dimeric H-NS contains two DNA-binding domains that can bridge two DNA duplexes, hence even though the concentration of H-NS is sufficient to cover only 1% of the genome during exponential growth, bridging of distant DNA sites may significantly affect DNA compaction (1). E. coli HU undoubtedly also plays a significant role in nucleoid organization [e.g. compare Supplementary Figure S3, panels A and B (2,6–8)], but recent evidence suggests that it may in fact counteract compaction by H-NS (57). While compaction by E. coli HU is seen at lower concentrations, where the observed compaction is attributed to DNA bending, higher protein concentrations were reported to induce rigid, helical filaments not associated with DNA condensation (56,58,59). Combined with the differential accessibility of DNA in complex with either TmHU or HBsu, we propose, therefore, that protein–DNA filaments produced at high concentrations of a given HU homolog are distinct, perhaps dictated by the type of networked protein–DNA interactions that are permissible based on the formation of either the flexible DNA bend that appears to go hand-in-hand with a shorter DNA site size or the large static bend associated with a longer occluded DNA site.
How does TmHU organize DNA into a compact network in which the DNA is largely buried? Three pieces of evidence contribute to an emerging model: (i) TmHU can wrap ∼37 bp of duplex, perhaps associated with a large DNA bend in which the DNA lies largely in a single plane; (ii) an apparent reduction in occluded site size with longer DNA [notably, this is distinct from measurements with Helicobacter pylori HU which was seen to occlude ∼20 bp, regardless of total DNA length (31)] and (iii) a DNase I cleavage pattern that suggests ordered placement of TmHU (Figure 7).
The modestly preferred binding of TmHU to DNA with appropriately spaced 4-nt loops (18) would place one TmHU protomer at the center of the 89 bp looped DNA (Figure 7). This is borne out by the DNase I cleavage pattern; on either side of the central cleavage sites at positions 50 and 42, which are separated by seven protected base pairs, are 9 and 11 protected sites, respectively, followed by cleavage sites separated by 8 bp. With the shorter protected segments likely corresponding to the sequence between intercalation sites, a model materializes in which TmHU protomers bind in alternating orientations such that DNA flanking the kinks is sandwiched between adjacent proteins (Figure 9). For the 89 bp looped DNA construct, three TmHU dimers would occupy the central ∼69 bp, leaving ∼10 bp at either end, insufficient for additional TmHU protomers. Notably, this model is reminiscent of the crystal packing of Anabaena HU–DNA complexes in which the shorter DNA substrates stack end-to-end (23). According to this model, DNA buried between adjacent TmHU protomers would be inaccessible to DNase I, as would DNA shielded by intercalating prolines at the tips of the β-strands. This mode of binding would explain the apparent reduction in occluded site size on increasing DNA length, the significant complex stability and the lack of staining by EtBr, and it is consistent with recent single molecule experiments which suggested close packing (60).
Figure 9.
Proposed model of DNA compaction by TmHU. Cartoon showing TmHU associating with long DNA, for which we propose that DNA flanking the sites of kinking becomes sandwiched between adjacent protomers. For association with the 89 bp looped DNA (Figure 7), the periodic cleavage pattern likely corresponds to positions of DNA intercalation (represented as 90° DNA angles in schematic).
In conclusion, TmHU appears capable of two distinct binding modes; it binds a 37 bp single site as an architectural factor, presumably introducing ∼160° bends, perhaps directed by preferred binding to the apex of a plectonemic superhelix. Accretion of TmHU protomers leads to constraint of negative supercoiling in the presence of topoisomerase I and to formation of a very compact association that affords significant DNA protection. The latter binding mode, which may be enhanced at the physiological growth temperature of T. maritima, would protect genomic DNA from damage or denaturation at elevated temperatures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the Socolofsky Microscopy Center, LSU, for support and assistance. This publication was also made possible by NIH Grant Number P20 RR16456 from the BRIN Program of the National Center for Research Resources and by a grant from the Howard Hughes Medical Institute through the Undergraduate Biological Sciences Education Program to Louisiana State University. This work was supported by National Science Foundation MCB-0414875 to A.G. The Open Access publication charges for this article were waived.
Conflict of interest statement. None declared.
REFERENCES
- 1.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from. Escherichia coli. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 2.Kellenberger E, Arnold-Schultz-Gahmen B. Chromatins of low-protein-content: special features of their compaction and condensation. FEMS Microbiol. Lett. 1992;100:361–370. doi: 10.1111/j.1574-6968.1992.tb14064.x. [DOI] [PubMed] [Google Scholar]
- 3.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 4.Achenbach-Richter L, Gupta R, Stetter KO, Woese CR. Were the original eubacteria thermophiles? Syst. Appl. Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- 5.Bocchetta M, Gribaldo S, Sanangelantoni A, Cammarano P. Phylogenetic depth of the bacterial genera Aquifex and Thermotoga inferred from analysis of ribosomal protein, elongation factor, and RNA polymerase subunit sequences. J. Mol. Evol. 2000;50:366–380. doi: 10.1007/s002399910040. [DOI] [PubMed] [Google Scholar]
- 6.Rouvière-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosome like structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 7.Griffith JD. Visualization of prokaryotic DNA in a regularly condensed chromatin-like fiber. Proc. Natl Acad. Sci. USA. 1976;73:563–567. doi: 10.1073/pnas.73.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunetti R, Prosseda G, Beghetto E, Colonna B, Micheli G. The looped domain organization of the nucleoid in histone-like protein defective Escherichia coli strains. Biochimie. 2001;83:873–882. doi: 10.1016/s0300-9084(01)01331-1. [DOI] [PubMed] [Google Scholar]
- 9.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 10.McGovern V, Higgins NP, Chiz RS, Jaworski A. H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie. 1994;76:1019–1029. doi: 10.1016/0300-9084(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 11.Kar S, Edgar R, Adhya S. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl Acad. Sci. USA. 2005;102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription. J. Biol. Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 13.Pontiggia A, Negri A, Beltrame M, Bianchi ME. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 14.Castaing B, Zelwer C, Laval J, Boiteux S. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 15.Balandina A, Kamashev D, Rouvière-Yaniv J. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 2002;277:27622–27628. doi: 10.1074/jbc.M201978200. [DOI] [PubMed] [Google Scholar]
- 16.Pinson V, Takahashi M, Rouvière-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 17.Esser D, Rudolph R, Jaenicke R, Bohm G. The HU protein from Thermotoga maritima: recombinant expression, purification and physicochemical characterization of an extremely hyperthermophilic DNA-binding protein. J. Mol. Biol. 1999;291:1135–1146. doi: 10.1006/jmbi.1999.3022. [DOI] [PubMed] [Google Scholar]
- 18.Grove A, Lim L. High-affinity DNA binding of HU protein from the hyperthermophile Thermotoga maritima. J. Mol. Biol. 2001;311:491–502. doi: 10.1006/jmbi.2001.4763. [DOI] [PubMed] [Google Scholar]
- 19.Bonnefoy E, Rouvière-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991;10:687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka I, Appelt K, Dijk J, White SW, Wilson S. 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984;310:376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- 21.Vis H, Mariani M, Vorgias CE, Wilson KS, Kaptein R, Boelens R. Solution structure of the HU protein from. Bacillus stearothermophius J. Mol. Biol. 1995;254:692–703. doi: 10.1006/jmbi.1995.0648. [DOI] [PubMed] [Google Scholar]
- 22.Rice PA, Yang S-W, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 23.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christodoulou E, Rypniewski WR, Vorgias CR. High-resolution X-ray structure of the DNA-binding protein HU from the hyper-thermophilic Thermotoga maritima and the determinants of its thermostability. Extremophiles. 2003;7:111–122. doi: 10.1007/s00792-002-0302-7. [DOI] [PubMed] [Google Scholar]
- 25.Mouw KW, Rice PA. Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol. Microbiol. 2007;63:1319–1330. doi: 10.1111/j.1365-2958.2007.05586.x. [DOI] [PubMed] [Google Scholar]
- 26.Grove A, Saavedra TC. The role of surface-exposed lysines in wrapping DNA about the bacterial histone-like protein HU. Biochemistry. 2002;41:7597–7603. doi: 10.1021/bi016095e. [DOI] [PubMed] [Google Scholar]
- 27.Kamau E, Tsihlis ND, Simmons LA, Grove A. Surface salt bridges modulate DNA site size of bacterial histone-like HU proteins. Biochem. J. 2005;250:49–55. doi: 10.1042/BJ20050274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broyles SS, Pettijohn DE. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 29.Le Hegarat F, Salti-Montesanto V, Hauck Y, Hirschbein L. Purification and characterization of the HU-like protein HPB9 from the Bacillus subtilis nucleoid. Biochim. Biophys. Acta. 1993;1172:101–107. doi: 10.1016/0167-4781(93)90275-i. [DOI] [PubMed] [Google Scholar]
- 30.Ross MA, Setlow P. The Bacillus subtilis HBsu protein modifies the effects of alpha/beta-type, small acid-soluble spore proteins on DNA. J. Bacteriol. 2000;182:1942–1948. doi: 10.1128/jb.182.7.1942-1948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Ghosh S, Grove A. Substrate specificity of Helicobacter pylori HU is determined by insufficient stabilization of DNA flexure points. Biochem. J. 2004;383:343–351. doi: 10.1042/BJ20040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobryn K, Naigamwalla DZ, Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 2000;37:145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 33.Bensaid A, Almeida A, Drlica K, Rouvière-Yaniv J. Cross-talk between topoisomerase I and HU in Escherichia coli. J. Mol. Biol. 1996;256:292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- 34.Guipaud O, Marguet E, Noll KM, Bouthier de la Tour C, Forterre P. Both DNA gyrase and reverse gyrase are present in the hyperthermophilic bacterium Thermotoga maritima. Proc. Natl Acad. Sci. USA. 1997;94:10606–10611. doi: 10.1073/pnas.94.20.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitehall SK, Kassavetis GA, Geiduschek EP. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh S, Grove A. Histone-like protein HU from Deinococcus radiodurans binds preferentially to four-way DNA junctions. J. Mol. Biol. 2004;337:561–571. doi: 10.1016/j.jmb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Wang JC. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J. Mol. Biol. 1974;89:783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- 38.Malcolm AD, Snounou G. Netropsin increases the linking number of DNA. Cold Spring Harb. Symp. Quant. Biol. 1983;47:323–326. doi: 10.1101/sqb.1983.047.01.037. [DOI] [PubMed] [Google Scholar]
- 39.Holbrook JA, Tsodikov OV, Saecker RM, Record MT., Jr Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 40.Grove A. Surface salt bridges modulate DNA wrapping by the type II DNA binding protein TF1. Biochemistry. 2003;42:8739–8747. doi: 10.1021/bi034551o. [DOI] [PubMed] [Google Scholar]
- 41.Rudnick J, Bruinsma R. DNA-protein cooperative binding through variable-range elastic coupling. Biophys. J. 1999;76:1725–1733. doi: 10.1016/S0006-3495(99)77334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köhler P, Marahiel MA. Association of the histone-like protein HBsu with the nucleoid of Bacillus subtilis. J. Bacteriol. 1997;179:2060–2064. doi: 10.1128/jb.179.6.2060-2064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- 44.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 45.Bouthier de la Tour C, Portemer C, Huber R, Forterre P, Duguet M. Reverse gyrase in thermophilic eubacteria. J. Bacteriol. 1991;173:3921–3923. doi: 10.1128/jb.173.12.3921-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Decanniere K, Babu AM, Sandman K, Reeve JN, Heinemann U. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol. 2000;303:35–47. doi: 10.1006/jmbi.2000.4104. [DOI] [PubMed] [Google Scholar]
- 47.Grayling RA, Sandman K, Reeve JN. DNA stability and DNA binding proteins. Adv. Protein Chem. 1996;48:437–467. doi: 10.1016/s0065-3233(08)60368-x. [DOI] [PubMed] [Google Scholar]
- 48.Marc F, Sandman K, Lurz R, Reeve JN. Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling. J. Biol. Chem. 2002;277:30879–30886. doi: 10.1074/jbc.M203674200. [DOI] [PubMed] [Google Scholar]
- 49.Musgrave DR, Sandman KM, Reeve JN. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc. Natl Acad. Sci. USA. 1991;88:10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamiche A, Carot V, Alilat M, De Lucia F, O’Donohue MF, Revet B, Prunell A. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: potential flipping of the protein from a left- to a right-handed superhelical form. Proc. Natl Acad. Sci. USA. 1996;93:7588–7593. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Napoli A, Kvaratskelia M, White MF, Rossi M, Ciaramella M. A novel member of the bacterial-archaeal regulator family is a nonspecific DNA-binding protein and induces positive supercoiling. J. Biol. Chem. 2001;276:10745–10752. doi: 10.1074/jbc.M010611200. [DOI] [PubMed] [Google Scholar]
- 52.Pavlicek JW, Oussatcheva EA, Sinden RR, Potaman VN, Sankey OF, Lyubchenko YL. Supercoiling-induced DNA bending. Biochemistry. 2004;43:10664–10668. doi: 10.1021/bi0362572. [DOI] [PubMed] [Google Scholar]
- 53.Pulleyblank DE, Shure M, Tang D, Vinograd J, Vosberg HP. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc. Natl Acad. Sci. USA. 1975;72:4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Depew DE, Wang JC. Conformational fluctuations of DNA helix. Proc. Natl Acad. Sci. USA. 1975;72:4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang JC. Helical repeat of DNA in solution. Proc. Natl Acad. Sci. USA. 1979;76:200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skoko D, Yan J, Johnson RC, Marko JF. Low-force DNA condensation and discontinuous high-force decondensation reveal a loop-stabilizing function of the protein Fis. Phys. Rev. Lett. 2005;95:208101. doi: 10.1103/PhysRevLett.95.208101. [DOI] [PubMed] [Google Scholar]
- 57.Dame RT, Goosen N. HU: promoting or counteracting DNA compaction? FEBS Lett. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 58.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sagi D, Friedman N, Vorgias C, Oppenheim AB, Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes. J. Mol. Biol. 2004;341:419–428. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 60.Salomo M, Kroy K, Kegler K, Gutsche C, Struhalla M, Reinmuth J, Skokov W, Immisch C, Hahn U, Kremer F. Binding of TmHU to single dsDNA as observed by optical tweezers. J. Mol. Biol. 2006;359:769–776. doi: 10.1016/j.jmb.2006.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.