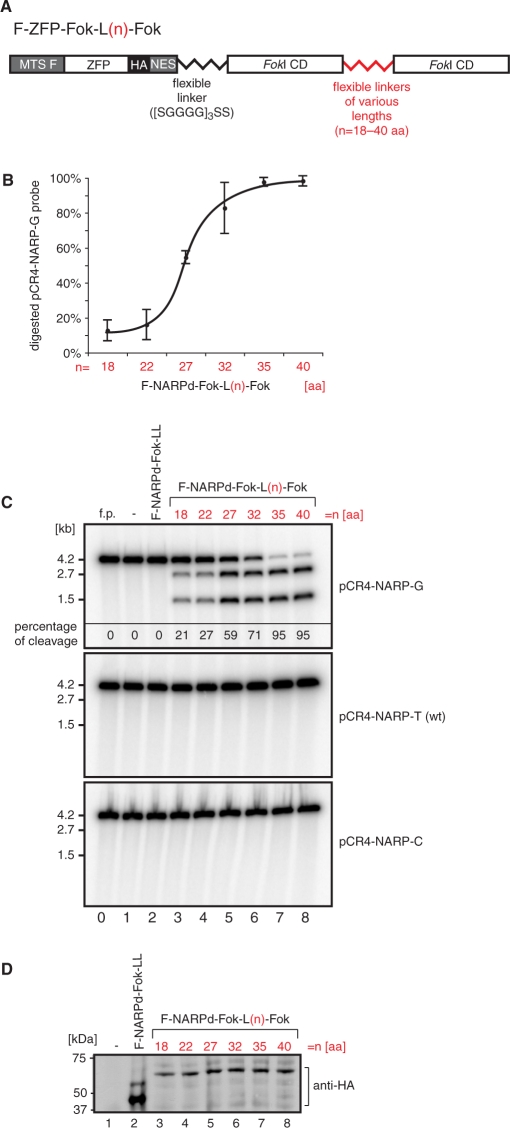
Figure 2.
Designing a single-chain ZFN to target a mitochondrial point mutation. (A) Schematic structure of mitochondrially targeted single-chain nucleases. The mitochondrial targeting sequence of F1β subunit of mitochondrial ATP synthase (MTS F) was fused to the N-terminus of the ZFP. The HA epitope tag and the NES facilitating mitochondrial targeting were added to the C-terminus of the ZFP. Two FokI domains were fused on the C-terminus of the ZFP and linked together via flexible linkers of various lengths denoted L(n), where n is the length of the linker in amino acids. (B) Optimization of the single-chain ZFN based on the NARPd construct (see Figure 1C). Variants of F-NARPd-Fok-L(n)-Fok were subjected to the in vitro assay as described in Materials and methods section using the specific target DNA in order to determine the optimal length of the linker between the two tethered FokI domains. The plot shows the results of the assay performed three times. (C) In vitro cleavage assay testing the specificity of the F-NARPd-Fok-L(n)-Fok constructs. The assay was performed as described in Materials and methods section. The following probes have been used: pCR4-NARP-G—containing the m.8993T>G substitution, pCR4-NARP-T —wt (i.e ‘T’) at the 8993 position and pCR4-NARP-C— containing the m.8993T>C substitution. The f.p denotes free probe. Specific digestion at the m.8993T>G mutation site results in the formation of 2.7- and 1.5-kb DNA fragments. The percentage of cleavage of the pCR4-NARP-G probe is given in the top panel. (D) Western blot illustrating that the constructs used in the test above are produced with the same efficiency in the in vitro transcription/translation system. ZFNs were detected with anti-HA antibody.