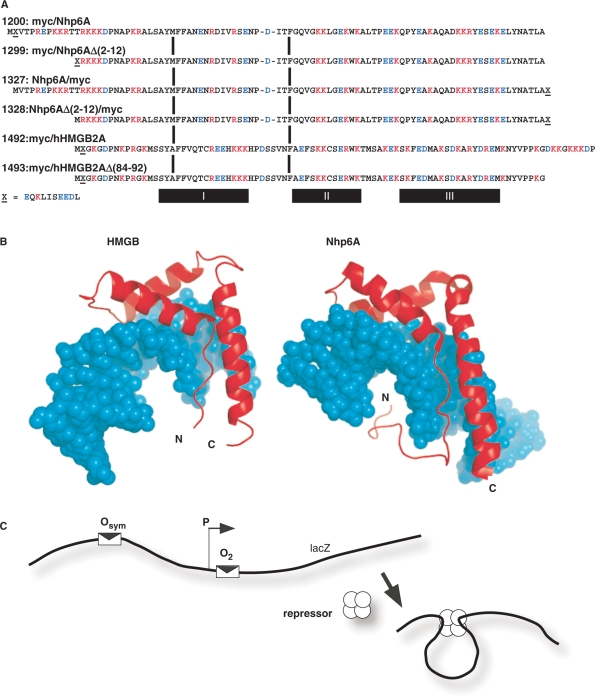
Figure 1.
Proteins tested in E. coli in vivo DNA-looping assay. (A) HMGB proteins tested. The plasmid number and a description of protein domains (amino-to-carboxyl order) are indicated, along with the complete amino acid sequence aligned at conserved HMG box intercalating residues (vertical bars). The c-Myc epitope tag is indicated by ‘X’ and its sequence is given below the figure. Deletion derivatives removed portions of cationic tails at the amino (constructs 1299, 1328) or carboxyl (construct 1493) termini. The α-helical domains I, II and III are indicated below the diagram. (B) Molecular models of single HMG boxes bound to DNA. Left, HMG box A from mammalian HMGB1 (37,38,41), PDB code 1CKT, which is highly similar to the corresponding box from HMGB2. Right, S. cerevisiae Nhp6A (40), pdb code 1J5N. The strongly bent DNA molecules are shown as space-filling models in cyan. HMGB domains are shown in red. Note that both the amino and carboxyl termini (N and C, respectively) are positioned so that basic tail extensions can neutralize crowded DNA backbone phosphates in the compressed major groove. (C) Schematic representation of episomal lac looping assay constructs (14,15). Weak (O2) and strong (Osym) lac operators are positioned at various separation distances flanking an E. coli promoter (P) upstream of the lacZ reporter gene encoding β-galactosidase. In the presence of tetrameric lac repressor protein, DNA looping causes repression of the test promoter. The dependence of DNA looping on operator separation in a series of strains where this distance is varied in base pair increments provides information about the apparent bending and torsional stiffness of DNA in vivo. When experiments are performed in ΔHU cells, the effect of apparent DNA flexibility of exogenous eukaryotic HMGB protein expression can be monitored.