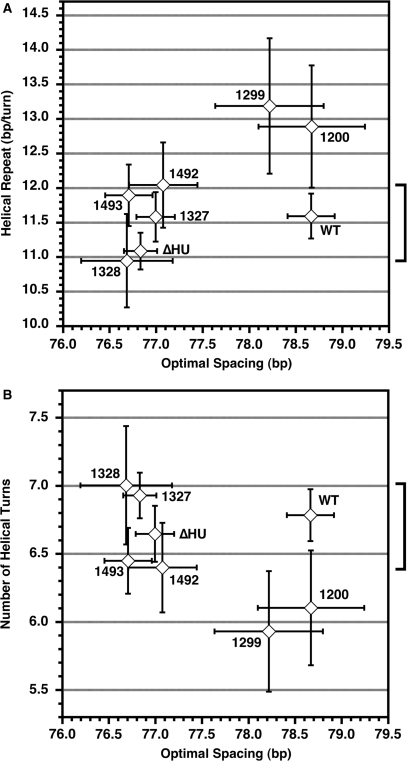
Figure 7.
Optimal spacing in terms of base pairs varies among bacterial strains but the optimal number of DNA helical repeats between operators is relatively consistent. (A) The range of helical repeat values from Table 2 (–IPTG only) versus the optimal loop spacing, with the bracket indicating that WT falls in the middle of the range of the well-determined helical repeats. (B) The number of helical turns between operators (calculated as optimal spacing/helical repeat) also clusters around the WT value. Error bars indicate 95% confidence limits on the parameters, with the estimated error in the number of helical turns being given by standard propagation of errors. The fitting parameters for constructs 1200 and 1299 are deemed unreliable, as discussed in the text.