Abstract
Actin-myosin II filament-based contractile structures in striated muscle, smooth muscle, and nonmuscle cells contain the actin filament-cross-linking protein α-actinin. In striated muscle Z-disks, α-actinin interacts with N-terminal domains of titin to provide a structural linkage crucial for the integrity of the sarcomere. We previously discovered a long titin isoform, originally smitin, hereafter sm-titin, in smooth muscle and demonstrated that native sm-titin interacts with C-terminal EF hand region and central rod R2-R3 spectrin-like repeat region sites in α-actinin. Reverse transcription-PCR analysis of RNA from human adult smooth muscles and cultured rat smooth muscle cells and Western blot analysis with a domain-specific antibody presented here revealed that sm-titin contains the titin gene-encoded Zq domain that may bind to the α-actinin R2-R3 central rod domain as well as Z-repeat domains that bind to the EF hand region. We investigated whether the sm-titin Zq domain binds to α-actinin R2 and R3 spectrin repeat-like domain loops that lie in proximity with two-fold symmetry on the surface of the central rod. Mutations in α-actinin R2 and R3 domain loop residues decreased interaction with expressed sm-titin Zq domain in glutathione S-transferase pull-down and solid phase binding assays. Alanine mutation of a region of the Zq domain with high propensity for α-helix formation decreased apparent Zq domain dimer formation and decreased Zq interaction with the α-actinin R2-R3 region in surface plasmon resonance assays. We present a model in which two sm-titin Zq domains interact with each other and with the two R2-R3 sites in the α-actinin central rod.
Smooth muscle cells use an extensively organized but poorly understood actin-myosin contractile apparatus to generate force for physiological requirements such as blood pressure regulation, airway constriction, and gut peristalsis. Dynamic changes in the organization of this contractile apparatus support a remarkable degree of smooth muscle cell mechanical and phenotypic plasticity in response to physiological cues (1, 2). Mechanical plasticity enables a smooth muscle cell to adapt to chronic or transient changes in cell length or load by reorganizing its contractile apparatus to maintain maximal force-generating capability (3, 4). Phenotypic plasticity permits a smooth muscle cell to convert reversibly from a “contractile” state, in which it can generate force for contraction, to a “synthetic” state, in which it can proliferate and migrate to participate in angiogenesis and wound healing or pathological formation of atherosclerotic plaques and thickening of arterial walls (5–8). Conversion between the contractile and synthetic phenotypic states involves changes in the expression and organization of smooth muscle contractile apparatus proteins (9). Some of these changes have been defined, but most of the underlying structural and molecular mechanisms supporting smooth muscle contractile apparatus organization and its plasticity have yet to be elucidated.
Although there are significant structural and stability differences, certain properties of the smooth muscle contractile apparatus organization resemble those of the better understood striated muscle sarcomere (10). Both striated and smooth muscle contractile systems contain antiparallel arrays of actin filaments, each of which anchors to one of a pair of α-actinin-containing functionally homologous structures, the striated muscle sarcomere Z-disk or the smooth muscle dense body. The myosin filaments, large bipolar filaments in striated muscles and large side-polar filaments in smooth muscle, interdigitate with antiparallel actin filaments and use similar crossbridge cycles of force production on actin filaments to pull the Z-disks and dense bodies closer together.
In striated muscles, a third major set of filaments, each of which is composed of a single molecule of the protein titin, organizes and stabilizes the sarcomere structure (11–14). Complete sequences of the human titin gene and of several differentially spliced titin gene transcripts expressed in vertebrate striated muscles are known (15). The longest striated muscle titin isoforms (∼1.0-μm-contour-length molecules composed of ∼28,000 amino acids) are elastic and span each half-sarcomere, with anchorage points in the Z-disk and M-line and on the myosin filaments. Titin filaments play important roles in sarcomere assembly, stability, and passive tension, as well as in stretch sensing, protein turnover, and gene regulation through interactions with other proteins including α-actinin (11, 16, 17).
We found a long isoform of the protein titin (originally named smitin, hereafter called sm-titin)3 in the smooth muscle contractile apparatus (18). The existence of titin isoforms in smooth muscle recently has been confirmed by others (19). We further demonstrated that sm-titin interacts in vitro with smooth muscle myosin (18) and smooth muscle α-actinin (20). It is likely that the sm-titin-α-actinin interaction plays major roles in establishing and maintaining the smooth muscle contractile apparatus and in regulating it dynamic organization. Determining the molecular basis for sm-titin-α-actinin interaction will yield valuable insight into this undoubtedly important linkage.
Smooth muscle α-actinin is localized in dense bodies, the functional equivalents of Z-discs in the smooth muscle contractile apparatus (21). The smooth muscle α-actinin is one of several isoforms of α-actinin encoded by a family of genes that are differentially expressed in different tissues (22). All α-actinins exist as rod-shaped antiparallel homodimers. Each monomer contains an actin-binding domain at the N terminus, four spectrin-like triple helical repeat regions (R1–4) in the central rod domain, and four C-terminal end cryptic calmodulin-like EF-hands (EF1–4), which in striated and smooth muscle isoforms fail to bind Ca2+.
In the striated muscle sarcomere, interaction with N-terminal domains of titin localizes α-actinin in the Z-disk and specifies the number of α-actinin cross-links between overlapping actin filaments (23). This key structural linkage is required for the assembly and structural integrity of the vertebrate striated muscle sarcomere (24). Disruption of this interaction by expression of a dominant-negative titin fragment causes sarcomere breakdown (25).
Two types of titin N-terminal domains interact with two different sites on α-actinin. One type of titin-α-actinin interaction involves binding any of seven differentially spliced 45-residue titin Z-repeat (Zr) domains to the EF34 set of cryptic EF-hand motifs in the C-terminal domain of each monomer on each end of the α-actinin antiparallel dimer (24). The structure of this interaction revealed how an α-helical region of the titin Zr7 domain lies in a groove formed by the EF34 domain in striated muscle α-actinin (26–28). We have confirmed that the sm-titin Zr7 domain interacts with similar cryptic EF hands near the C terminus of smooth muscle α-actinin, but regulation of this interaction appears to be different in striated and smooth muscles (20).
A second type of interaction occurs between the striated muscle titin Zq domain and the second and third (R2-R3) spectrin-like triple helical repeats in the striated muscle α-actinin rod (24). The molecular basis for this interaction remains very poorly characterized, even in the interaction between striated muscle titins and striated muscle α-actinin, in part because the entire R2-R3 region appears to be required for Zq binding. Here, we report characterization and molecular mapping of a similar interaction between the sm-titin Zq domain and the R2-R3 region of the smooth muscle α-actinin central rod (24). Based on evidence presented here, we propose a model for how Zq domain interaction with loop regions of both R2 and R3 and with itself may contribute to the sm-titin-α-actinin linkage in smooth muscle.
EXPERIMENTAL PROCEDURES
Smooth Muscle RT-PCR Analysis—RT-PCR analysis of commercially available human carotid artery, bladder, and uterus RNAs (Stratagene) and Trizol-isolated RNA from the A7r5 rat aorta smooth muscle cell line was used to determine whether a smooth muscle titin isoform contained known α-actinin-binding domains and the sequence of that region. Primers complementary to human and rat titin gene sequence encoding this region were designed using PrimerQuest (Integrated DNA Technologies, Coralville, IA). First strand cDNA was synthesized using SuperScript III (Invitrogen) and primers complementary to the sense strand of exons 20 and 28. Subsequent PCR amplification was done using forward and reverse primer pairs covering sequences in the 5′ end of human titin exons 1, 8, and 20 (forward) and the 3′ end of exons 14, 20, and 28 (reverse). The standard PCR protocol used included 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 0.5 °C below the lowest primer calculated melting temperature, and extension for 1 min/kb of possible product sequence at 68 °C. The length of products was assessed when compared with standards on an agarose gel and purified using a gel extraction kit (Qiagen, Valencia, CA). Purified products were sequenced in both directions using the PCR amplification primers and analyzed for exon-exon junctions to ensure mRNA origin. Forward primers used were designed from the sequences of exon 1, 5′CGTTTCAGAAGCAACCTTGGGCTT-3′; exon 8, 5′ACTGCTGTGCACATCCAACCTGCT-3′; and exon 20, 5′ACCTGCCGCGCCTTACTTTATTAC-3′. The reverse primers used were from exon 14, 5′TGACTGCTTTAGGGACAACGTGGG-3′; exon 20, 5′CCTGCTTGTTCCTCTGTGAGGCTA-3′; and exon 28, 5′TACCAGTTGACTTTGGGCTGAGGGT-3′.
Fusion Protein Expression and Mutagenesis—The human sm-titin Zq domain was amplified from human carotid RNA using primers designed against the 5′ end of titin exon 15 and the 3′ end of titin exon 16, cloned into pET-15b vector (Invitrogen), and expressed as a C-terminal His6-tagged fusion protein in BL-21 (DE3) Escherichia coli. After nickel-nitrilotriacetic acid agarose (Pierce) affinity chromatography, FPLC size exclusion chromatography through Superdex 75 (GE Healthcare) run at a rate of 0.5 ml/min was used to remove minor contaminants. The human smooth muscle α-actinin spectrin-like repeat domain (ACTN1-R2-R3-GST) was cloned and expressed as a GST fusion protein, as described previously (20). Alanine and phosphomimetic mutations of α-actinin R2-R3 loop (K421A, Y423A, T425A, T427D, N586A, T590A, and N591A) and sm-titin Zq domain (S43A/Q53A/K64A) residues were obtained by site-directed mutagenesis with the Stratagene QuikChange site-directed mutagenesis kit.
For surface plasmon resonance (SPR) experiments, purified ACTN1-R2-R3-GST was cleaved with thrombin (10 units, Sigma) according to manufacturer's recommendations and passed over immobilized glutathione agarose (Pierce) to isolate the ACTN1-R2-R3 domain. Purified WT-Zq and the S43A/Q53A/K64A-Zq triple mutant were purified to homogeneity (at least 95% pure, as determined by SDS-PAGE) and dialyzed extensively into HBS-EP+ running buffer (Biacore Life Sciences, Piscataway, NJ). Protein concentrations were calculated using predicted extinction coefficients (ProtParam site at us.expasy.org), and absorbance at 280 nm was obtained from a full wavelength scan using a Varian Cary 300 UV/VIS spectrophotometer.
Gel Electrophoresis and Western Blots of Native sm-Titin— Chicken striated pectoralis muscle and pig aorta and uterus tissues were obtained from local slaughterhouses. The tissues where transported to the laboratory on ice, minced, quick-frozen in liquid nitrogen, and pulverized into powder. The powder was extracted using 1× SDS-sample buffer. The SDS extract was fractionated on highly porous 4–20% gradient SDS-polyacrylamide, as described previously (29). The gel was electroblotted onto nitrocellulose using semidry blot transfer system (Bio-Rad Laboratories) and stained using Ponceau S. The blots were blocked in 5.0% nonfat dry milk for 1–2 h at room temperature. The anti-TKZ rabbit polyclonal primary antibody raised in the T. Keller laboratory against an expressed titin region containing the Z-repeats Zr1, Zr2, Zr3, and Zr7 and the Zq domain, as described previously (30), was applied to the membrane and incubated for 1–2 h at room temperature. The membrane then was washed extensively in TBS-Tween 20 and incubated for 1 h with a secondary antibody conjugated to horseradish peroxidase. The chemiluminescent signal was developed using the Amersham Biosciences ECL Plus Western blotting detection system (according to the manufacturer's recommendations).
Immunolocalization of sm-Titin in Cryosections and Cultured Cells—A segment of porcine aorta obtained from a slaughterhouse was placed into ice-cold cryoprotectant solution containing sucrose-phosphate and fixed overnight in a solution containing freshly prepared 3.7% formaldehyde and 0.1% Triton-X-100. The frozen aorta was cross-sectioned at a thickness of 10 μm in a cryostat. The sections were collected on 2% gelatin-coated slides, and the cryosection slides were incubated with anti-TKZ polyclonal antibody (diluted 1:100) followed by Alexa Fluor 488-coupled secondary antibody (diluted 1:500), mounted with ProLong® Gold mounting medium (Invitrogen), and viewed with a Zeiss LSM 510 (Zeiss, Inc., Thornwood, NY).
The A7r5 rat aorta and human coronary artery smooth muscle cells were grown on Nafion®-coated coverslips for 3–4 days (31). The cells were fixed with 3.7% formaldehyde in 1× phosphate-buffered saline for 15 min and then permeabilized with 0.2% Triton X-100-phosphate-buffered saline for 15 min at room temperature. To minimize nonspecific binding, the coverslips were blocked in 10% goat serum, 0.05% Triton X-100-phosphate-buffered saline for 60 min before incubation at 37 °C for 1 h each in the anti-TKZ polyclonal antibody primary (diluted 1:100) and Alexa Fluor 488-conjugated (diluted 1:300) secondary antibodies. The A7r5 cells were double-labeled with phalloidin (rhodamine) and the anti-TKZ polyclonal antibody. The human coronary artery cells were labeled with only the anti-TKZ polyclonal antibody. These coverslips were observed with a Leica TCS SP2 AOBS laser confocal microscope.
ACTN1-R2-R2-GST Binding Assays—ACTN1-R2-R3-GST or mutations were purified to homogeneity and immobilized onto glutathione-agarose beads (1 mg/ml). Purified sm-titin Zq domain (0.5 mg/ml) was dialyzed into interaction buffer containing 100 mm KCl and 0.2% Triton X-100 and mixed with the ACTN1-R2-R3-GST beads overnight at 4 °C. The beads were pelleted by centrifugation, washed twice with interaction buffer, and eluted with 1× SDS-PAGE sample buffer. The amount of sm-titin Zq binding was assessed by SDS-PAGE of the elutant. All pull-down experiments were done in duplicate.
Solid-phase protein binding assay using microtiter plates was performed as described previously (20). For these experiments, microtiter plate wells were coated with expressed sm-titin Zq domain and incubated with ACTN1-R2-R2-GST in 100 mm KCl, 0.2% Triton X-100. The amount of ACTN1-R2-R2-GST bound was assessed with anti-GST antibody (1:1000; Affinity BioReagents, Golden, CO). The mean value and standard error were calculated from three separate experiments.
Surface Plasmon Resonance Experiments—For these experiments, 2000 resonance units of ACTN1-R2-R3 domain in 10 mm acetate pH 4.5 were immobilized on a Series S sensor chip CM5 (Biacore Life Sciences) using amine coupling chemistry. The sm-titin Zq domain (WT-Zq) or mutant Zq domain (S43A/Q53A/K64A-Zq) was dialyzed extensively into HBS-EP+ running buffer (Biacore Life Sciences), flowed over the chip at a rate of 10 μl/min (300 s of contact time), and regenerated with running buffer containing 1 m NaCl (30 μl/min). To determine steady state affinity, binding of various concentrations (0–12.8 μm) of WT-Zq or S43A/Q53A/K64A-Zq were assayed. Binding constants were extrapolated from response (resonance units) versus time (seconds) values using the Biacore Life Sciences T-100 analysis software. For relative ranking studies, WT-Zq and S43A/Q53A/K64A-Zq were included in the same experiment, and binding was compared directly from the resulting sensorgrams. All sensorgrams were double zeroed with values of the running buffer alone and from a reference cell consisting of activated/blocked surface and no ACTN1-R2-R3 domain. All experiments were done in duplicate using a Biacore Life Sciences T-100 system housed in the Florida State University College of Medicine Biomedical Proteomics Laboratory.
Graphic Modeling and Sequence Alignment—The Visual Molecular Dynamic viewer (University of Illinois at Urbana-Champaign) was used for theoretical modeling of the mutations in the loops of the α-actinin R2-R3 domain and modeling of the Zq domain binding to this region (Protein Data Bank 1SJJ). Sequence similarity and alignment were produced using ClustalW European Molecular Biology Laboratory-European Bioinformatics Institute) and Jalview (Barton Group, University of Dundee).
RESULTS
Smooth Muscle RNA Contains a Titin Transcript Encoding Z-repeat and Zq α-Actinin-binding Domains—RT-PCR analysis of commercially available human carotid artery, uterus, and bladder total RNAs yielded cDNAs with sequences matching those of exons from the exon 1–28 region of the only known human titin gene. All cDNAs obtained lacked intron sequence and crossed predicted exon-exon boundaries, proving they arose from mRNA transcripts and not contaminating genomic DNA. The three smooth muscle RNA sources each yielded a single cDNA product (∼2100 bp) from a pair of primers designed to exon 1 and exon 14 of the titin gene that was significantly smaller than the predicted size of the product if all exon sequences were present. Sequence analysis verified the exclusion of exons 11–13, with all products bridging the exon boundary between exons 10 and 14 (Fig. 1). The excluded exons 11–13 encode the Z-repeats 4–6. The cDNA products obtained therefore encode the Z1 and Z2 Ig domains, the Zis1 unique sequence, and the α-actinin-binding domains known as the Z-repeats Zr1, Zr2, Zr3, and Zr7 and the Zq domain. The cDNA products obtained using primers designed to exons 20 and exon 28 contained the sequences of all known intervening exons (Fig. 1B). We found the same pattern of exon usage by similar RT-PCR analysis of total RNA isolated from rat aortic smooth muscle A7r5 cells grown in culture.
FIGURE 1.
Smooth muscle titin RT-PCR products. A, the diagram of the 28 exons comprising the 5′ end of the human titin gene (modified from Bang et al. (15)) illustrates exons included in RT-PCR products amplified from three commercially available RNA sources (human carotid artery, uterus, and bladder RNA) and RNA isolated from cultured rat aorta smooth muscle A7r5 cells. The light gray boxes encode Ig domains. The dark gray boxes encode unique domains. The hatched box exons encode the seven Z-repeats and the Zq domain (exon 14) that interact with α-actinin. The hatched boxes in the shaded area indicate Z-repeat exons encoding Zr4–6 not found in the PCR products. The arrows (not to scale) designate positions of the exon 1, 8, and 20 forward primers (right arrows) and the exon 14, 20, and 28 reverse primers (left arrows) used for the RT-PCR analysis. UTR, untranslated region. B, the relationship between the possible PCR product size if all exon sequences are included and the actual PCR product size obtained reveals the exclusion of the Z-repeat 4, 5, and 6 encoding exons 11, 12, and 13 covered by the shaded box.
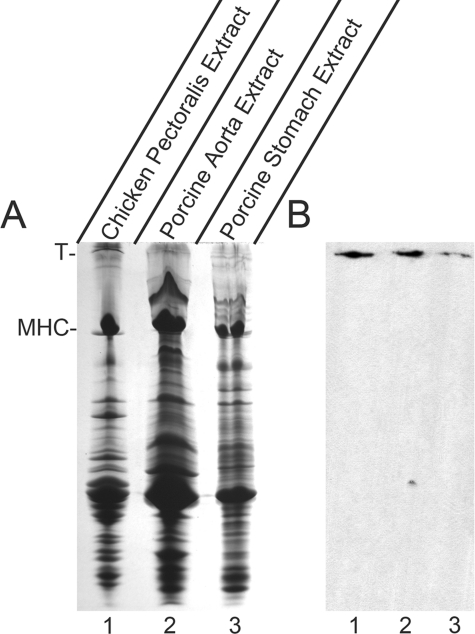
Western Blot Demonstrates Presence of sm-Titin Protein in Smooth Muscle—To confirm the expression of titin containing the α-actinin-binding region in adult smooth muscle tissues, we raised a rabbit polyclonal antibody (anti-TKZ) against the region encoded by cloned exons 8–14 expressed as a GST fusion protein in bacteria. Western blot analysis using the anti-TKZ antibody demonstrated a reactive band in crude extracts of porcine aorta and stomach smooth muscle (Fig. 2B, lanes 2 and 3) that migrated at a rate similar to that of chicken skeletal muscle titin (Fig. 2B, lane 1). Lack of reactivity with any of the other bands in the crude extracts demonstrated the high titin specificity of the anti-TKZ antibody. Reactivity of the anti-TKZ antibody, which was raised against an expressed fragment encoded by titin gene sequence, with the protein previously identified by the laboratory as smitin confirms smitin as a smooth muscle isoform of titin. Henceforth, we will refer to this protein as sm-titin instead of smitin.
FIGURE 2.
Anti-TKZ antibody reacts with the high molecular weight sm-titin band in smooth muscle extracts. The anti-TKZ rabbit polyclonal antibody was raised against a bacterially expressed fragment composed of the sm-titin α-actinin-binding region (Z-repeats Zr1, Zr2, Zr3, and Zr7 and the Zq domain). A, SDS-PAGE gel stained with Coomassie Blue of SDS extracts of chicken pectoralis striated muscle used as a standard for migration of titin (lane 1) and two porcine smooth muscles (lanes 2 and 3). T, titin; MHC, myosin heavy chain. B, Western analysis using anti-TKZ antibody of a blot containing duplicate samples run on the same gel and electroblotted to nitrocellulose. In each smooth muscle extract (lanes 2 and 3), the anti-TKZ antibody reacts with a single antibody-positive band that migrates at a rate similar to the titin in the chicken pectoralis striated muscle extract (lane 1).
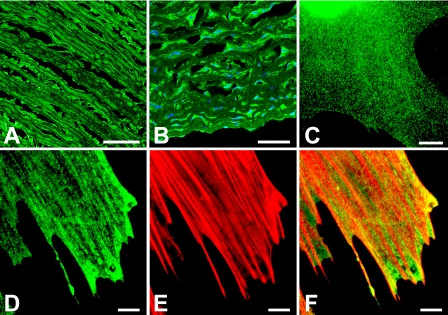
Sm-Titin Immunolocalization—Immunofluorescence localization using the anti-TKZ antibody demonstrated the presence of sm-titin in porcine aorta and cultured smooth muscle cells (Fig. 3). In intact porcine aorta, anti-TKZ labeled the smooth muscle layer of the tissue and not the elastic filaments lining the aorta (Fig. 3, A and B). In cultured smooth muscle cells, the anti-TKZ antibody stained in a punctate pattern along the stress fiber-like contractile apparatus structures (Fig. 3, C–F).
FIGURE 3.
Anti-TKZ antibody immunofluorescent localization of the Z-repeat-Zq region of smooth muscle titin in cryosections of smooth muscle and cultured smooth muscle cells. A and B, cells in the smooth muscle layers of 10-μm-thick cryo cross-sections of adult porcine aorta were stained with anti-TKZ sm-titin primary antibody detected with Alexa Fluor 488-labeled secondary antibody (green) and 4′,6-diamidino-2-phenylindole to label the nuclei. The sm-titin antibody stained the cells in the smooth muscle layers but not the layers of elastic filaments between the layers of cells. C, anti-TKZ antibody staining of human coronary artery smooth muscle cells and rat aorta smooth muscle A7r5 cells were grown for 4 days on a polyelectrolyte multilayer (Nafion®)-coated coverslip. Growth on Nafion induces the cells to become contractile. The anti-smtitin antibody (D) yields linear, punctate patterns of labeling aligned with but not completely overlapping rhodamine phalloidin-stained actin filaments (E). F, merged image of D and E. Bar in A, 500 μm; bar in B, 250 μm; bars in C–F, 20 μm.
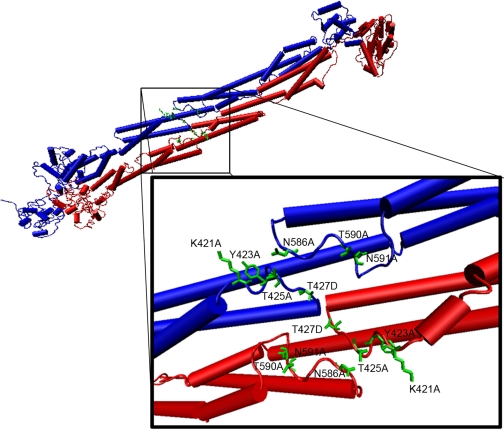
Loops in the R2-R3 Domain Mediate sm-Titin Binding—In reviewing a recent model of smooth muscle α-actinin structure (32), it became apparent that loops from the spectrin-repeat R2 and R3 domains could constitute the unknown binding site for the sm-titin Zq domain. The structural model predicts that an R2 domain loop (residues 421–425) and an R3 domain loop (residues 586–591), which are 145 residues apart in the primary sequence, lie in proximity on the surface of the α-actinin monomer rod and that the sites from both monomers lie on the same side of the dimer (Fig. 4). The juxtaposition of these loops forms an apparent groove on the surface of the a-actinin rod that we hypothesized could constitute the binding site for a helical sm-titin Zq domain.
FIGURE 4.
Model representation of the smooth muscle α-actinin central rod spectrin repeat-like R2 and R3 domains and key loop residues. A structural model of the smooth muscle α-actinin R2 and R3 domains in the central rod (Liu et al. (32)) was modified to show positions of the R2 and R3 loop residues mutated for sm-titin Zq binding interaction experiments. One monomer peptide of the antiparallel α-actinin homodimer is shown in blue. The antiparallel monomer is shown in red. Sites formed by the R2 and R3 loops in each monomer lie in proximity with a two-fold symmetry on the same side of the α-actinin central rod.
We used pull-down experiments with alanine mutations of several R2 loop residues (K421A, Y424A, and T425A) and R3 loop residues (N586A, T590A, and N591A), and a phosphomimetic mutation of an R2 loop threonine residue (T427D) in a putative protein kinase CKII phosphorylation site (Thr-427-lsE), to investigate whether the predicted α-actinin R2 and R3 loops contribute to the site that binds the titin Zq domain. For the GST pull-down experiments, the R2-R3 domain region constructs were expressed as GST fusion proteins, and the titin Zq region was expressed as a His-tagged fragment. Glutathione-coupled beads efficiently pelleted the GST-R2-R3 domain alone (Fig. 5A, lane 6). A saturable amount of the Zq domain co-pelleted when bound to the wild type R2-R3 (Fig. 5A, lane 5), but none pelleted in the absence of the R2-R3 domain under the same conditions (Fig. 5B, lane 4). Alanine mutations of several residues from both the R2 loop domain and the R3 loop domain significantly decreased Zq binding indicated by the amount of Zq co-pelleted with the R2-R3 region (Fig. 5C).
FIGURE 5.
Sm-titin Zq domain binding to wild type and mutated smooth muscle α-actinin R2-R3 domains in vitro. A–C, wild type and mutant smooth muscle α-actinin R2-R3 domains expressed as GST fusion proteins and immobilized on glutathione beads were incubated with an expressed wild type smooth muscle Zq domain fragment in buffer containing 100 mm KCl and 0.2% Triton X-100. T, titin; MHC, myosin heavy chain. A, SDS-PAGE gel from a representative experiment shows unbound Zq domain in the supernatant and in washes (lanes 2–4), Zq domain that bound to the α-actinin R2-R3 domain on the beads and coeluted with SDS (lane 5), and R2-R3 fragment alone bound to and eluted from the beads (lane 6). Lane 1 contains a chicken pectoralis muscle extract used as a molecular weight standard. B, a control experiment demonstrates that the Zq domain fails to bind to beads lacking the α-actinin R2-R3 domain (lane 4). C, the Zq region of a gel showing the amount of bound Zq (equivalent to A, lane 5) from experiments in which R2-R3 domains with the specified mutations were bound to the beads reveals variable effects of the mutations on Zq binding. The mutations tested include the R2 loop mutations K421A, Y423A, and T425A, the R3 loop mutations N586A, T590A, N591A, and the phosphomimetic mutation T427D of a putative CK2 phosphorylation site. The signs indicate the lanes in which Zq binding was detectable (+) or undetectable (-) on the gel.
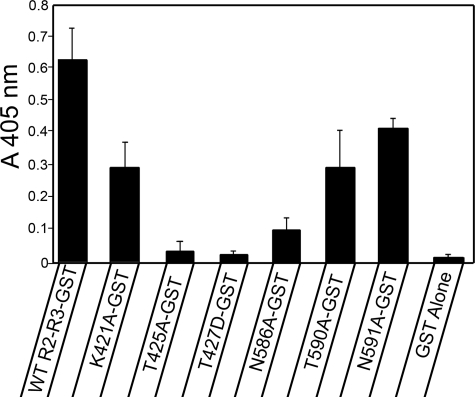
A solid phase binding assay, which revealed relative amounts of the R2-R3 domain wild type and mutant fragments bound to immobilized wild type titin Zq domain, confirmed the results of the pull-down assay. All of the R2-R3 domain mutants exhibited a significant decrease in binding when compared with the wild type binding, with those exhibiting the least interaction in the pull-down assay also yielding the least binding in the solid phase assay (Fig. 6). It is interesting to note that alanine mutation of both the R2-Thr-425 and the R3-Asn-586, which the model predicts to be in proximity, has significantly less effect on binding than mutation of each residue alone (data not shown).
FIGURE 6.
Solid phase binding assay confirms mutation of α-actinin R2-R3 loop residues causes decrease in sm-titin Zq binding. Microtiter wells coated with the expressed sm-titin Zq domain were incubated with WT and mutated α-actinin R2-R3 GST fusion proteins or GST alone. The presence of GST was detected with an anti-GST antibody followed by a secondary antibody conjugated to alkaline phosphatase and incubation with substrate for color development. The A405 nm mean ± S.E. for each mutant was calculated from three separate experiments.
Taken together, these results support the hypothesis that loops from the R2 and R3 domains combine to form a binding site for the titin Zq domain. Moreover, the effect of the phosphomimetic mutation of the R2 domain Thr-427 residue (T427D), which is part of a predicted casein kinase II phosphorylation site (TlsE), raises the possibility that phosphorylation may regulate this region of sm-titin-α-actinin interaction, although evidence for phosphorylation of the rod region of α-actinin in vivo remains lacking.
A Helical Region in the sm-Titin Zq Domain Is Important for α-Actinin Binding—We used SPR to quantify sm-titin Zq domain interaction with wild type smooth muscle α-actinin R2-R3 fragment and to identify the region of sm-titin Zq that interacts with the α-actinin. Wild type sm-titin Zq domain bound to immobilized α-actinin R2-R3 domain with a KD of 344 ± 3 nm (Fig. 7).
FIGURE 7.
Sm-titin Zq domain binds to α-actinin R2-R3 domain with nanomolar affinity. A, the surface plasmon resonance responses were recorded for various wild type sm-titin Zq concentrations (0–6400 nm) flowed at 10 μl/min over wild type α-actinin R2-R3 domain that was immobilized on a Biacore Life Sciences CM5 sensor chip. RU, resonance units. B, the binding curve generated using mean sensorgram plateau resonance units values from two experiments indicates that the wild type smooth muscle Zq domain binds to the wild type α-actinin with a KD = 344 ± 3 nm.
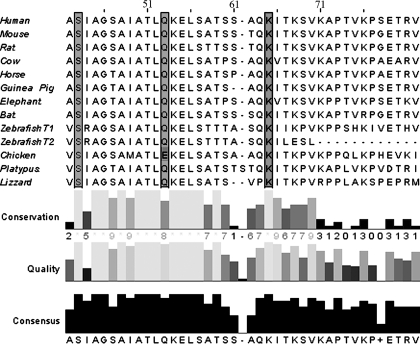
Site-directed alanine mutagenesis of Zq domain residues identified the segment that interacts with the R2-R3 domain to a highly conserved, mostly helical, region. The titin Zq domain fragment used for all the α-actinin R2-R3 binding studies described contains 135 amino acids encoded by sequence in titin gene exons 15 and 16. A region of this sequence is highly conserved across a wide variety of vertebrate species (Fig. 8). Several secondary structure prediction programs applied to the entire 135-residue human sequence predict various amounts of extended β-structure (14–40%), random coil (49–56%), and α-helix (11–30%), but all support a high propensity for α-helix formation in the highly conserved region of the titin Zq sequence. Reasoning that a highly conserved α-helical Zq region may bind in the groove between the α-actinin R2 and R3 domain loops, we used SPR and pulldown experiments to test the effect of alanine mutation of three well conserved residues in the predicted Zq domain. In the SPR analysis, the triple S43A/Q53A/K64A-Zq mutant bound very poorly to the immobilized α-actinin R2-R3 domain when compared with wild type Zq in relative rank studies (Fig. 9A). For these experiments, equal concentrations of the wild type and mutant Zq domains were tested, but only one wild type concentration that exhibits binding in the range of that of the highest mutant concentrations is shown in the sensorgram. Subsequent steady state affinity experiments using the Zq triple mutant failed to reach saturation even at the highest Zq domain concentration (data not shown). Each of the two Zq single mutations that yielded soluble expressed protein, the S43A and the K64A mutations, also significantly inhibited binding to the α-actinin R2-R3 domain in pulldown experiments (Fig. 9B). These results suggest that the highly conserved helical region in the Zq domain may bind to the conserved surface loops from the R2 and R3 spectrin repeat-like domains.
FIGURE 8.
The predicted sm-titin Zq domain contains highly conserved residues. ClustalW alignment of the sm-titin Zq domain predicted from the titin gene sequences of representative vertebrates reveals highly conserved regions. The highly conserved residues Ser-43, Gln-53, and Lys-64 (highlighted) each were mutated to alanine. The numbering reflects residue position in the primary sequence encoded by human titin gene exons 15 and 16.
FIGURE 9.
Mutation of the sm-titin Zq domain significantly decreased affinity for the α-actinin R2-R3 domain. A, a surface plasmon resonance relative ranking experiment was used to compare the binding of wild type Zq domain (WT-Zq) domain and the S43A/Q53A/K64A-Zq triple mutant (SQK-Zq). An expanded region of the sensorgram generated from binding of one wild type concentration (156 nm) and two triple mutant concentrations (625 and 1250 nm) demonstrated a significantly (more than 8-fold) weaker binding of the mutant when compared with the wild type. RU, resonance units. B, a pulldown assay using a WT GST-α-actinin R2-R3 domain bound to glutathione beads pelleted the WT-Zq domain but failed to pellet either of the Zq domains with the single S43A and K64A single mutations.
A gel filtration step in the protocol routinely used to purify each expressed fragment revealed one possible difference between the wild type Zq and the mutant Zq domains. The wild type Zq domain, with a calculated molecular mass of ∼17 kDa, eluted through a Superdex 75 gel filtration FPLC column as a single peak (Fig. 10A) containing a single protein band that migrates in SDS-PAGE at a rate consistent with a 17-kDa peptide (Fig. 10B). In contrast, the SQK-Zq triple mutant eluted through the gel filtration column as two peaks (Fig. 10A), both of which contain only the 17-kDa protein on SDS-PAGE (Fig. 10C). Comparison of the elution rates of the single WT-Zq peak and the leading SQK-Zq peak with the elution rates of globular protein standards reveled that these peaks had apparent molecular masses (∼40 kDa) greater than twice that of the 17 kDa predicted for the expressed Zq peptide. The elution rate of the trailing SQK-Zq peak when compared with those of the standards predicts a protein with a molecular mass of ∼27 kDa. These results are most consistent with the possibilities that in solution, the WT-Zq forms an elongate dimer and the SQK triple mutation destabilizes the dimer to yield pools of dimers and elongate monomers, each of which elute at greater than expected rates because of their elongate shapes.
FIGURE 10.
FPLC size exclusion chromatography of bacterially expressed wild type and triple mutant Zq domains on Superdex 75. Superimposed Superdex 75 FPLC elution profiles (A) of the wild type Zq domain (WT-Zq) and the S43A/Q53A/K64A-Zq triple mutant (SQK-Zq) and SDS-PAGE of the column fractions (B and C) demonstrates the WT-Zq migrates as a single peak (A, solid line) containing the predicted ∼17.5 Zq peptide (B), whereas the SQK-Zq migrates as two peaks (A, dashed line), each of which contains the ∼17.5 Zq peptide (C). Based on migration rates of protein standards run on the same column, the migration rates of the single WT-Zq peak and the leading SQK-Zq peak are predictive of a protein with a molecular mass of ∼40 kDa, which is consistent with an elongate Zq dimer, and the migration rate of the trailing SQK-Zq peak is predictive of a protein with a molecular mass of ∼27 kDa, which is consistent with an elongate Zq monomer. The slight shift in peak fractions between the scan (A) and gels (B and C) reflects the volume in the column system between the scan and fraction collection sites. The Superdex 75 column was standardized with bovine serum albumin (66 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), RNase A (14 kDa), and cytochrome c (12 kDa). The 24 kDa and the 15 kDa indicate the migration positions of molecular mass standards (stds MW) on the SDS gels. Abs, absorbance.
DISCUSSION
We previously mapped smooth muscle α-actinin-binding sites for native sm-titin to the central rod R2-R3 spectrin-like repeats and C-terminal domains, sites that are analogous to those in striated muscle α-actinin that bind respectively to striated muscle titin Zq and Z-repeat domains (20). Whether Z-repeat and Zq domains are present in any adult smooth muscle titin isoforms recently was brought into doubt, however, by an investigation that revealed the presence of smooth muscle transcripts containing sequence from only 80–90 titin gene exons and excluding the Z-repeat- and Zq-encoding exons (19). We used RT-PCR analysis and antibody detection to confirm the presence of these domains in the long adult smooth muscle titin isoform that we originally characterized as smitin (18, 33) and now call sm-titin. RT-PCR analysis of human carotid artery RNA yielded products encoding Z-repeats Zr1, Zr2, Zr3, and Zr7 and the Zq domain. We also found these domains in RT-PCR products amplified from human bladder and uterus RNA, as well as from RNA isolated from cultured rat aortic smooth muscle A7r5 cells (data not shown). Other titin regions encoded by RT-PCR products from these RNAs include the largest exon of the myosin-binding domain, the C-terminal kinase domain, and uniquely spliced portions of the PEVK domain (data not shown).
In addition, an antibody (anti-TKZ) that was raised against the Z-repeat-Zq region and reacts with striated muscle titin also reacted in Western blots with the very high molecular protein originally characterized by us as smitin. The anti-TKZ antibody also yielded strong immunofluorescence signal from smooth muscle cells in cryosections of adult smooth muscle. Moreover, the antibody revealed a punctate pattern of epitope localization along the stress fiber-like structures in the contractile apparatus of A7r5 cells that were induced to become contractile by culture on a polyelectrolyte substrate.4
Inspiration for this investigation arose from visual analysis of a smooth muscle α-actinin structural model (32), which revealed that loops from the R2 and R3 spectrin-like domains lie in proximity to each other on the rod surface, despite being separated by ∼150 residues in the primary sequence. The model also indicates that identical R2-R3 loop pairs form a two-fold axis of symmetry on the same side of the α-actinin antiparallel homodimer rod domain. Based on this model, we targeted certain R2-R3 loops residues to determine whether alanine mutagenesis would have any effect on Zq domain binding. We found that single alanine mutations of residues in the R2-R3 domain loops region significantly reduced sm-titin Zq domain binding in GST pulldown experiments and solid phase binding assays. Moreover, a phosphomimetic mutation of the R2 domain Thr-427 residue (T427D) also decreased binding (Fig. 5C), raising the possibility that phosphorylation may regulate sm-titin-α-actinin interaction. Although such phosphorylation has yet to be demonstrated in vivo, it may occur only transiently during structural rearrangement of the contractile apparatus for mechanical or phenotypic plasticity of the smooth muscle cell.
The specific region of the Zq domain that interacts with the R2-R3 loops was suggested by existing deletion analysis of cardiac titin and cardiac α-actinin constructs (24, 34) and analysis presented here demonstrating a high degree of sequence conservation across various vertebrate titins in a region predicted to have high α-helical propensity. Using SPR analysis, we confirmed that a triple alanine mutation of this region significantly decreased Zq domain affinity for immobilized α-actinin R2-R3 rod. Various algorithms predict that the region encompassed by the triple mutation (SIAGSAIATLQKELSATSSAQK; bold residues were mutated to alanine for SQK triple mutant) forms an amphipathic helix or perhaps a broken helix, which might be optimal for lying in the groove between the α-actinin R2 and R3 loops. Considering that in sm-titin, the more N-terminal Zr7 domain interacts with the α-actinin C-terminal EF34, it is reasonable to predict that the N-terminal end of the interacting Zq sequence is oriented toward the Thr-427–Thr-591 end of the R2-R3 groove on the same monomer.
The proximity on the same side of the rod and the two-fold symmetry of the R2-R3 loop alignments supports the possibility that Zq helical regions lying in the R2-R3 loop grooves might interact with each other in an antiparallel orientation. Interestingly, although purifying the bacterially expressed wild type Zq domain with size-exclusion FPLC chromatography, we found it eluted as if it had twice the anticipated molecular weight, perhaps indicating dimerization. In contrast, the triple mutant yielded two elution peaks: the apparent dimer peak and a slower peak migrating at the rate predicted for a 17-kDa monomer. Confirmation of antiparallel interaction of helices in this region of the Zq domain will require additional experimentation.
Based on these results and our predicted orientation of the Zq domain interaction, we present a model of sm-titin Zq domain interaction with the smooth muscle α-actinin rod domain in which Zq domains from two sm-titins homodimerize to promote and support the protein complex (Fig. 11). If correct, this demonstrates how a single α-actinin molecule could cross-link two sm-titin molecules in the smooth muscle dense body independently of other possible titin cross-linking proteins, such as Telethonin/Tcap (35–37), which have yet to be identified in the smooth muscle contractile apparatus.
FIGURE 11.
Model for titin Zq domain interaction with the α-actinin R2-R3 central rod domain. This model proposes that loops in the R2 and R3 spectrin repeat-like domains in each of the antiparallel monomers, which lie in proximity on the surface of the central rod with a two-fold symmetry, constitute the binding site for the 26-residue helical region of the Zq domain (yellow). Proximity of two Zq domains interacting with the R2-R3 domain may allow antiparallel interaction of the helical regions of the Zq domains, which may promote stability of the complex.
Acknowledgments
We thank other members of the T. Keller laboratory for advice and encouragement, Margaret Seavy for help with protein purification, Scott Olenych and Rani Dhanarajan for help with molecular cloning, Kim Riddle for help with immunofluorescence microcopy, XiXi Jia for help with ultrathin cryosections, and Maroun Moussallem and Dr. J. B. Schlenoff for help with the Polyelectrolyte Multilayer surfaces.
This work was supported, in whole or in part, by National Institutes of Health Grant R01EB006158 (to J. Schlenoff (PI) and T. K. (co-PI)). This work was also funded by Grant 017297 from the Florida State University Cornerstone Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: sm-titin, smooth muscle titin; RT-PCR, reverse transcription-PCR; FPLC, fast protein liquid chromatography; GST, glutathione S-transferase; WT, wild type; SPR, surface plasmon resonance; Zr, Z-repeat.
S. G. Olenych, M. D. Mousallem, S. L. Scott, A. F. Miller, J. B. Schlenoff, and T. C. S. Keller III, manuscript in preparation.
References
- 1.Gerthoffer, W. T., and Gunst, S. J. (2001) J. Appl. Physiol. 91 963-972 [DOI] [PubMed] [Google Scholar]
- 2.Seow, C. Y., Pratusevich, V. R., and Ford, L. E. (2000) J. Appl. Physiol. 89 869-876 [DOI] [PubMed] [Google Scholar]
- 3.Ford, L. E., Seow, C. Y., and Pratusevich, V. R. (1994) Can. J. Physiol. Pharmacol. 72 1320-1324 [DOI] [PubMed] [Google Scholar]
- 4.Seow, C. Y. (2000) J. Appl. Physiol. 89 2065-2072 [DOI] [PubMed] [Google Scholar]
- 5.Nakamura, A., Isoyama, S., Watanabe, T., Katoh, M., and Sawai, T. (1998) Microvasc. Res. 55 14-28 [DOI] [PubMed] [Google Scholar]
- 6.Newby, A. C., and Zaltsman, A. B. (2000) J. Pathol. 190 300-309 [DOI] [PubMed] [Google Scholar]
- 7.Orford, J. L., Selwyn, A. P., Ganz, P., Popma, J. J., and Rogers, C. (2000) Am. J. Cardiol. 86 6H-11H [DOI] [PubMed] [Google Scholar]
- 8.Zanellato, A. M. C., Borrione, A. C., Tonello, M., Scannapieco, G., Pauletto, P., and Sartore, S. (1990) Arteriosclerosis 10 996-1009 [DOI] [PubMed] [Google Scholar]
- 9.Halayko, A. J., and Solway, J. (2001) J. Appl. Physiol. 90 358-368 [DOI] [PubMed] [Google Scholar]
- 10.Herrera, A. M., McParland, B. E., Bienkowska, A., Tait, R., Pare, P. D., and Seow, C. Y. (2005) J. Cell Sci. 118 2381-2392 [DOI] [PubMed] [Google Scholar]
- 11.Linke, W. A. (2007) Cardiovasc. Res. 77 637-648 [DOI] [PubMed] [Google Scholar]
- 12.Clark, K. A., McElhinny, A. S., Beckerle, M. C., and Gregorio, C. C. (2002) Annu. Rev. Cell Dev. Biol. 18 637-706 [DOI] [PubMed] [Google Scholar]
- 13.Sanger, J. W., Ayoob, J. C., Chowrashi, P., Zurawski, D., and Sanger, J. M. (2000) Adv. Exp. Med. Biol. 481 89-102 [DOI] [PubMed] [Google Scholar]
- 14.Granzier, H. L., and Labeit, S. (2005) Adv. Protein Chem. 71 89-119 [DOI] [PubMed] [Google Scholar]
- 15.Bang, M. L., Centner, T., Fornoff, F., Geach, A. J., Gotthardt, M., McNabb, M., Witt, C. C., Labeit, D., Gregorio, C. C., Granzier, H., and Labeit, S. (2001) Circ. Res. 89 1065-1072 [DOI] [PubMed] [Google Scholar]
- 16.Lange, S., Xiang, F., Yakovenko, A., Vihola, A., Hackman, P., Rostkova, E., Kristensen, J., Brandmeier, B., Franzen, G., Hedberg, B., Gunnarsson, L. G., Hughes, S. M., Marchand, S., Sejersen, T., Richard, I., Edstrom, L., Ehler, E., Udd, B., and Gautel, M. (2005) Science 308 1599-1603 [DOI] [PubMed] [Google Scholar]
- 17.Tskhovrebova, L., and Trinick, J. (2005) Nat. Med. 11 478-479 [DOI] [PubMed] [Google Scholar]
- 18.Kim, K., and Keller, T. C., III (2002) J. Cell Biol. 156 101-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labeit, S., Lahmers, S., Burkart, C., Fong, C., McNabb, M., Witt, S., Witt, C., Labeit, D., and Granzier, H. (2006) J. Mol. Biol. 362 664-681 [DOI] [PubMed] [Google Scholar]
- 20.Chi, R. J., Olenych, S. G., Kim, K., and Keller, T. C., III (2005) Int. J. Biochem. Cell Biol. 37 1470-1482 [DOI] [PubMed] [Google Scholar]
- 21.Small, J. V., and Gimona, M. (1998) Acta Physiol. Scand. 164 341-348 [DOI] [PubMed] [Google Scholar]
- 22.Dixson, J. D., Forstner, M. J., and Garcia, D. M. (2003) J. Mol. Evol. 56 1-10 [DOI] [PubMed] [Google Scholar]
- 23.Sorimachi, H., Freiburg, A., Kolmerer, B., Ishiura, S., Stier, G., Gregorio, C. C., Labeit, D., Linke, W. A., Suzuki, K., and Labeit, S. (1997) J. Mol. Biol. 270 688-695 [DOI] [PubMed] [Google Scholar]
- 24.Young, P., Ferguson, C., Banuelos, S., and Gautel, M. (1998) EMBO. J. 17 1614-1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayoob, J. C., Turnacioglu, K. K., Mittal, B., Sanger, J. M., and Sanger, J. W. (2000) Cell Motil. Cytoskeleton 45 67-82 [DOI] [PubMed] [Google Scholar]
- 26.Joseph, C., Stier, G., O'Brien, R., Politou, A. S., Atkinson, R. A., Bianco, A., Ladbury, J. E., Martin, S. R., and Pastore, A. (2001) Biochemistry 40 4957-4965 [DOI] [PubMed] [Google Scholar]
- 27.Atkinson, R. A., Joseph, C., Kelly, G., Muskett, F. W., Frenkiel, T. A., Nietlispach, D., and Pastore, A. (2001) Nat. Struct. Biol. 8 853-857 [DOI] [PubMed] [Google Scholar]
- 28.Atkinson, R. A., Joseph, C., Dal Piaz, F., Birolo, L., Stier, G., Pucci, P., and Pastore, A. (2000) Biochemistry 39 5255-5264 [DOI] [PubMed] [Google Scholar]
- 29.Eilertsen, K. J., and Keller, T. C. S., III (1992) J. Cell Biol. 119 549-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavnar, P. J., Olenych, S. G., and Keller, T. C., III (2007) Cell Motil. Cytoskeleton 64 418-433 [DOI] [PubMed] [Google Scholar]
- 31.Salloum, D. S., Olenych, S. G., Keller, T. C., and Schlenoff, J. B. (2005) Biomacromolecules 6 161-167 [DOI] [PubMed] [Google Scholar]
- 32.Liu, J., Taylor, D. W., and Taylor, K. A. (2004) J. Mol. Biol. 338 115-125 [DOI] [PubMed] [Google Scholar]
- 33.Keller, T. C., III, Eilertsen, K., Higginbotham, M., Kazmierski, S., Kim, K. T., and Velichkova, M. (2000) Adv. Exp. Med. Biol. 481 265-277 [DOI] [PubMed] [Google Scholar]
- 34.Young, P., and Gautel, M. (2000) EMBO J. 19 6331-6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faulkner, G., Lanfranchi, G., and Valle, G. (2001) IUBMB Life 51 275-282 [DOI] [PubMed] [Google Scholar]
- 36.Lee, E. H., Gao, M., Pinotsis, N., Wilmanns, M., and Schulten, K. (2006) Structure (Camb.) 14 497-509 [DOI] [PubMed] [Google Scholar]
- 37.Gregorio, C. C., Trombitas, K., Centner, T., Kolmerer, B., Stier, G., Kunke, K., Suzuki, K., Obermayr, F., Herrmann, B., Granzier, H., Sorimachi, H., and Labeit, S. (1998) J. Cell Biol. 143 1013-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]