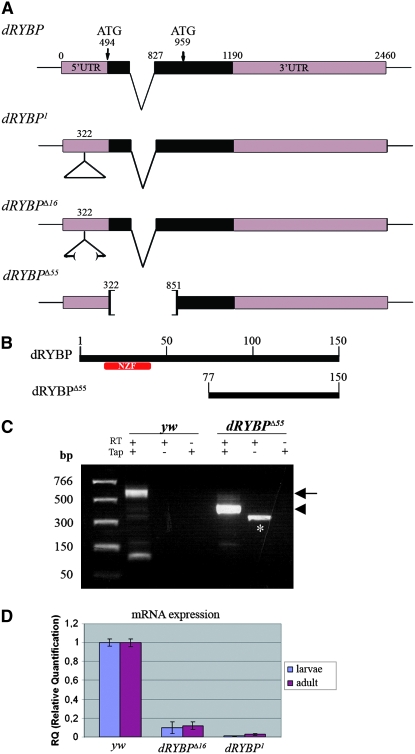
Figure 1.—
Mutations in the dRYBP gene, 5′-RACE and quantitative PCR analysis. (A) The dRYBP gene structure, showing the positions of the two in-frame ATGs codons. dRYBP1 contains the P[KG08683]-element insertion (indicated as a triangle) at nucleotide position 322. dRYBPΔ16 is an incomplete deletion of the P[KG08683] element. dRYBPΔ55 deletes nucleotides 322–853. (B) The dRYBP protein contains an NZF domain (red) located in the amino terminus. The putative dRYBPΔ55 protein encoded by dRYBPΔ55 does not contain the NZF domain (red). (C) 5′-RACE analysis was performed using total RNA isolated from homozygous Df(1)w67c23 (yw) and homozygous dRYBPΔ55 embryos. The reverse-transcription (RT) reaction was performed in the presence and in the absence of Tobacco acid pyrophosphatase (Tap). A control containing “minus-Tap”-treated RNA was also performed. A band of 560 bp (arrow) was observed, corresponding to the dRYBP wt 5′-RACE product (lane yw, “plus-RT/plus-Tap”; the smaller band in this lane is an artifact). A band of 380 bp (arrowhead) corresponds to the dRYBPΔ55 5′-RACE product (lane dRYBPΔ55, plus-RT/plus-Tap). The band in the second lane of dRYBPΔ55 (marked with *) corresponds to “plus-RT/minus-Tap” is an artifact of the 5′-RACE protocol used (Instructions manual, FirstChoice-RLM-RACE; Ambion). (D) Quantitative PCR results showing the relative mRNA expression levels from homozygous Df(1)w67c23 (yw), dRYBP1, and dRYBPΔ16 early first-instar larvae and adults.