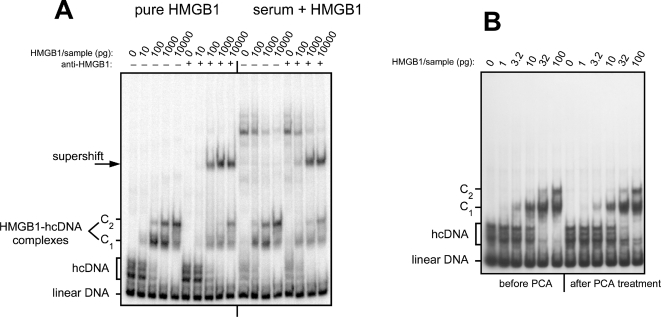
Figure 3. Detection of HMGB1 in human serum.
(A) On the left half of the gel, complexes obtained with increasing amounts of pure HMGB1: 0 pg, 10 pg, 100 pg, 1 ng, 10 ng (competitor DNA: 2 µg double strand plus 1 µg single strand per sample). The next five samples contain the same amounts of HMGB1 plus an anti-HMGB1 antibody, showing the supershift of the specific complexes (since pure HMGB1 was used here, and as antibodies were polyclonal and present in excess, the partial supershifting is most likely due to a low or moderate affinity of part of the antibodies). On the right-hand side of the gel, the same experiment was performed with the same amounts of HMGB1 and competitor DNA mixed with 1 µL of serum from a healthy individual. The overall pattern of shifted and supershifted bands is identical, over a background that contains a number of non-specific extra bands, despite the use of a large amount of nonspecific competitor DNA. (B) Purification of HMGB1 by PCA treatment. The left-hand part of the gel shows the interactions of hcDNA with the indicated amounts of pure HMGB1, from 1 pg to 100 pg. On the right-hand part of the gel, samples containing the same concentrations of HMGB1, plus 50 mg/mL BSA to mimic an HMGB1-free serum, were treated with PCA and neutralized as described in Materials and Methods, then assayed for hcDNA binding. Note that the hcDNA-binding activity is recovered without any loss after the PCA treatment procedure.