Abstract
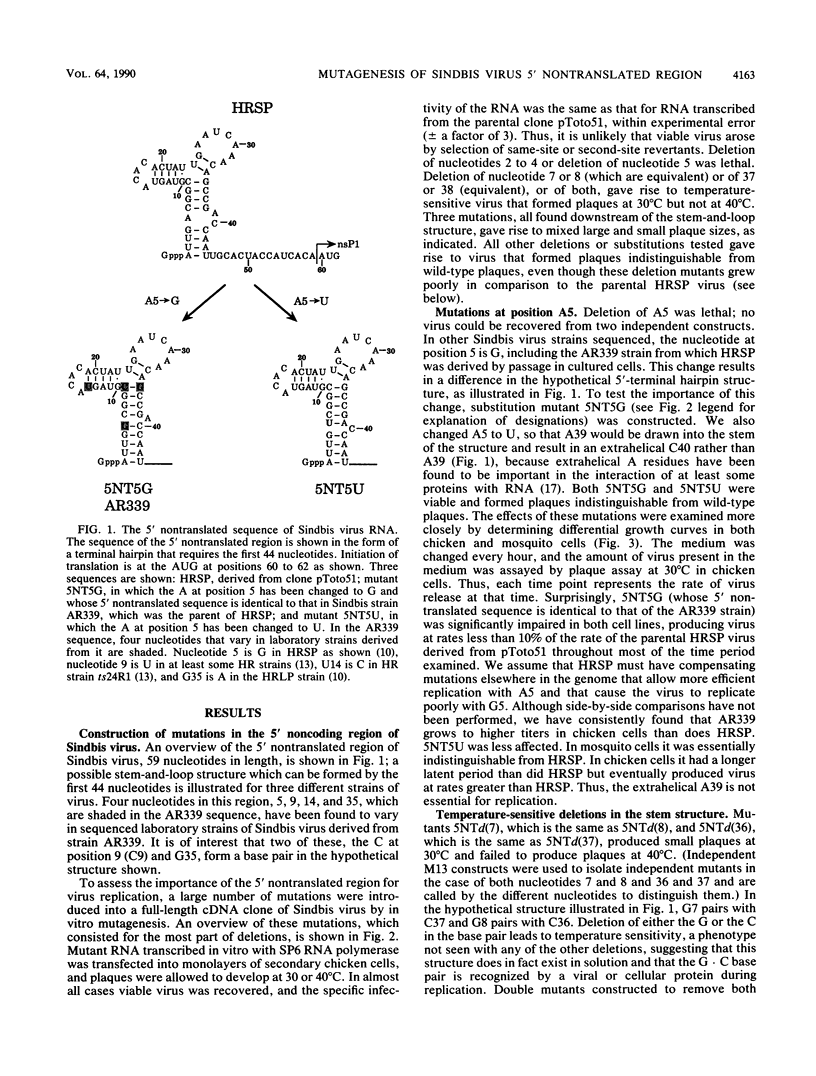
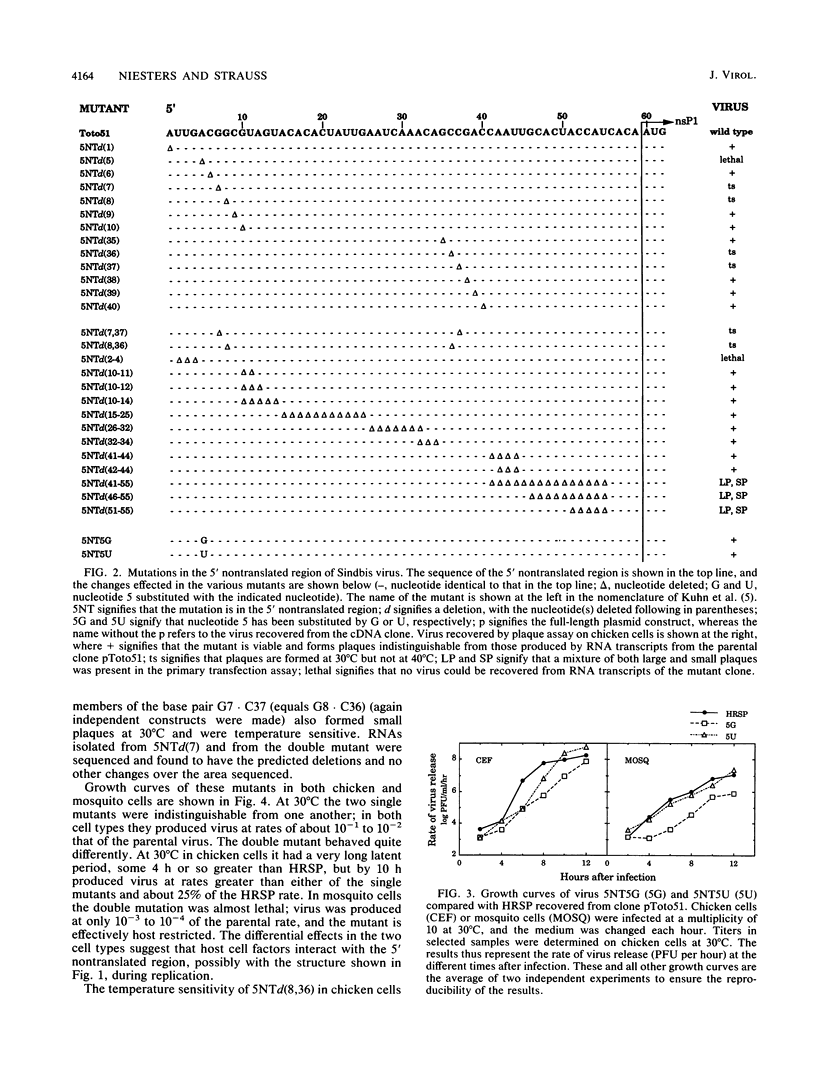
We have constructed 24 deletion mutants which contain deletions of from 1 to 15 nucleotides in the 5' nontranslated region of Sindbis virus RNA and tested the effect of these mutations on virus replication. The results showed that the first 44 nucleotides, which are capable of forming a hairpin structure, are important for virus replication, as all deletions tested in this region were either lethal or resulted in virus that grew poorly in comparison to the parental virus. Many of these deletions had different effects in mosquito cells than in chicken cells, suggesting that cellular factors, presumably proteins, bind to this region. This domain may function in at least two processes in viral replication. It seems likely that in the minus strand, this sequence element is bound by the viral replicase and promotes RNA replication. In the plus strand, this element may modulate initiation of translation of the nonstructural proteins. The results suggest that the hairpin structure itself is important. All deletions within it had deleterious effects on virus replication, and in particular, deletion of one of the G residues at nucleotide 7 or 8 or of one of the C residues at nucleotide 36 or 37 which are theoretically base-paired with these G's resulted in temperature-sensitive viruses that behaved very similarly. In contrast, large deletions between the 44-nucleotide hairpin and the translation start site at nucleotides 60 to 62 resulted in virus that grew as well as or better than the parental virus in both chicken and mosquito cells. The A residue at position 5 of the HRSP strain used was examined in more detail. Deletion of this A was lethal, whereas substitution by G resulted in a virus that grew poorly, despite the fact that G is present at position 5 in the AR339 parent of HRSP. U at position 5 resulted in a virus that grew less well than the A5 strain but better than the G5 mutant.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Levis R., Raju R., Huang H. V., Rice C. M. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J Virol. 1989 Dec;63(12):5216–5227. doi: 10.1128/jvi.63.12.5216-5227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J Virol. 1988 Mar;62(3):998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. J., Hong Z., Strauss J. H. Mutagenesis of the 3' nontranslated region of Sindbis virus RNA. J Virol. 1990 Apr;64(4):1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Schlesinger S., Huang H. V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990 Apr;64(4):1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B. G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986 Jan 17;44(1):137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Niesters H. G., Strauss J. H. Mutagenesis of the conserved 51-nucleotide region of Sindbis virus. J Virol. 1990 Apr;64(4):1639–1647. doi: 10.1128/jvi.64.4.1639-1647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Rice C. M., Dalgarno L., Strauss E. G., Strauss J. H. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Strauss E. G., Strauss J. H. The 5'-terminal sequences of the genomic RNAs of several alphaviruses. J Mol Biol. 1983 Jul 25;168(1):1–15. doi: 10.1016/s0022-2836(83)80319-2. [DOI] [PubMed] [Google Scholar]
- Ou J. H., Trent D. W., Strauss J. H. The 3'-non-coding regions of alphavirus RNAs contain repeating sequences. J Mol Biol. 1982 Apr 25;156(4):719–730. doi: 10.1016/0022-2836(82)90138-3. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D., Barkhimer D. B., Sawicki S. G., Rice C. M., Schlesinger S. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology. 1990 Jan;174(1):43–52. doi: 10.1016/0042-6822(90)90052-s. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Tsiang M., Weiss B. G., Schlesinger S. Effects of 5'-terminal modifications on the biological activity of defective interfering RNAs of Sindbis virus. J Virol. 1988 Jan;62(1):47–53. doi: 10.1128/jvi.62.1.47-53.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Uhlenbeck O. C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987 Dec 15;26(25):8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


