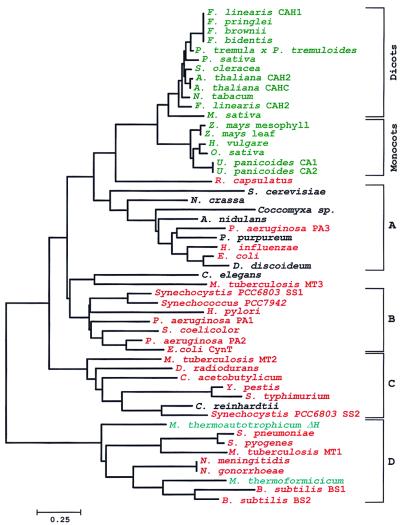
Figure 1.
Phylogeny of the β class of carbonic anhydrase. A neighbor-joining tree of proteins related to β carbonic anhydrases was constructed. Putative β carbonic anhydrases were identified in the Eukarya [plant sequences (green) and lower eukaryotes (black)], Bacteria (red), and Archaea (blue) domains. GenBank accession numbers or sequence source, their percent identity, and similarity to the M. thermoautotrophicum ΔH Cab (accession number 1272331) appear in parentheses, as follows, in alphabetical order: Arabidopsis thaliana CAH2 (1168739, 28.4, 58.0), Arabidopsis thaliana CAHC (115470, 26.9, 55.7), Aspergillus nidulans (AA786939, 18.0, 45.3), Bacillus subtilis BS1 (1945660, 40.4, 60.8), Bacillus subtilis BS2 (2293156, 39.0, 59.0), Caenorhabditis elegans (1049368, 32.5, 61.4), Chlamydomonas reinhardtii (1323549, 25.0, 53.8), Clostridium acetobutylicum (Genome Therapeutics, 27.2, 53.3), Coccomyxa sp. (1663720, 24.1, 50.6), Deinococcus radiodurans (The Institute for Genomic Research, 28.0, 53.6), Dictyostelium discoideum (1513236, 31.1, 56.3), Escherichia coli CynT (1657535, 34.2, 54.4), Escherichia coli (1723190, 29.0, 54.0), Flaveria bidentis (1168745, 27.5, 53.3), Flaveria brownii (1168746, 27.5, 53.3), Flaveria linearis CAH1 (1168738, 27.5, 56.3), Flaveria linearis CAH2 (882243, 27.8, 56.3), Flaveria pringlei (1168747, 27.5, 53.3), Haemophilus influenzae (1175500, 28.7, 57.3), Helicobacter pylori (2313081, 31.9, 53.4), Hordeum vulgare (729003, 25.3, 52.5), Medicago sativa (1938227, 26.0, 52.1), Methanobacterium thermoautotrophicum ΔH (1272331), Methanobacterium thermoformicicum (1279772, 51.8, 73.8), Mycobacterium tuberculosis MT1 (1722951, 34.4, 60.0), Mycobacterium tuberculosis MT2 (2950411, 26.6, 49.1), Mycobacterium tuberculosis MT3 (1877328, 28.4, 50.3), Neisseria gonorrhoeae (Univ. of Oklahoma Genome Center, 39.4, 64.7), Neisseria meningitidis (Sanger Centre, 39.4, 64.7), Neurospora crassa (AA901623, 29.8, 49.6), Nicotiana tabacum (115473, 25.0, 51.2), Oryza sativa (606817, 24.8, 53.3), Pisum sativum (115471, 22.6, 49.4), Populus tremula × Populus tremuloides (1354517, 23.3, 51.5), Porphyridium purpureum (1395172, 26.7, 55.8), Pseudomonas aeruginosa PA1 (Univ. of Washington/PathoGenesis, 31.5, 55.2), Pseudomonas aeruginosa PA2 (Univ. of Washington/PathoGenesis, 37.0, 59.3), Pseudomonas aeruginosa PA3 (Univ. of Washington/PathoGenesis, 29.6, 54.3), Rhodobacter capsulatus (Univ. of Chicago/Institute of Molecular Genetics, 27.4, 54.2), Saccharomyces cerevisiae Nce3p (1277232, 29.8, 48.2), Salmonella typhimurium (2460259, 28.7, 54.8), Spinacia oleracea (115472, 23.3, 52.7), Streptococcus pneumoniae (The Institute for Genomic Research, 34.8, 62.6), Streptococcus pyogenes (Univ. of Oklahoma Genome Center, 31.7, 62.7), Streptomyces coelicolor (Sanger Centre, 28.7, 52.1), Synechococcus PCC7942 (118069, 28.7, 56.9), Synechocystis PCC6803 SS1 (1653251, 30.9, 59.4), Synechocystis PCC6803 SS2 (1001130, 23.3, 51.5), Urochloa panicoides CA1 (882248, 20.7, 52.7), Urochloa panicoides CA2 (882246, 20.7, 52.7), Yersinia pestis (Sanger Centre, 28.1, 53.8), Zea mays mesophyll (606815, 23.0, 55.2), and Zea mays leaf (606811, 23.0, 55.6).