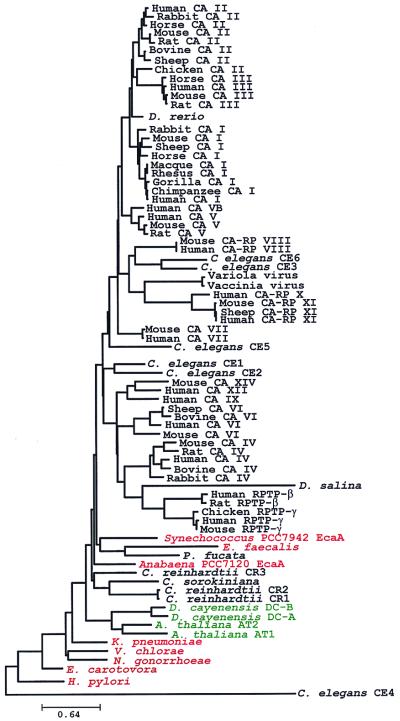
Figure 3.
Phylogeny of the α class of carbonic anhydrase. A neighbor-joining tree of proteins related to α carbonic anhydrases was constructed. Putative α carbonic anhydrase were identified in the Eukarya [plant sequences (green) and other eukaryotes (black)] and the Bacteria (denoted in red) domain. GenBank accession nos. or sequence source and their percent identity and similarity to the Anabaena PCC7120 α carbonic anhydrase (accession no. 1825485) appear in parentheses, as follows: Arabidopsis thaliana AT1 (1916014, 29.8, 39.3), Arabidopsis thaliana AT2 (AAD29832, 28.7, 38.0), bovine CA II (115453, 37.8, 48.3), bovine CA IV (1575298, 26.1, 35.3), bovine CA VI (1526572, 32.5, 38.3), Caenorhabditis elegans CE1 (1109858, 34.5, 42.3), Caenorhabditis elegans CE2 (2493489, 31.6, 39.1), Caenorhabditis elegans CE3 (746517, 27.2,35.3), Caenorhabditis elegans CE4 (CAA92190, 31.7, 36.5), Caenorhabditis elegans CE5 (AAA20616, 23.1, 41.1), Caenorhabditis elegans CE6 (3217404, 25.2, 34.1), chicken CA II (115454, 34.5, 41.6), chicken receptor-type tyrosine phosphatase γ (AAB16910, 27.3, 35.1), chimpanzee CA I (478302, 40.7, 47.3), Chlamydomonas reinhardtii CR1 (115447, 24.4, 32.4), Chlamydomonas reinhardtii CR2 (115455, 23.1, 30.7), Chlamydomonas reinhardtii CR3 (2301259, 37.0, 44.4), Chlorella sorokiniana (3133261, 17.4, 26.1), Danio rerio (3123190, 38.2, 45.3), Dioscorea cayenensis DCA (1364059, 30.6, 38.3), Dioscorea cayenensis DCB (2147328, 32.2, 40.8), Dunaliella salina (1431878, 26.7, 34.0), Enterococcus faecalis (The Institute for Genomic Research, 30.2, 35.7), Erwinia carotovora (2773324, 39.5, 50.6), gorilla CA I (478303, 41.2, 46.9), Helicobacter pylori (2314346, 23.4, 32.3), horse CA I (115448, 40.7, 46.9), horse CA II (223999, 37.5, 45.5), horse CA III (115461, 32.3, 45.1), human CA I (115449, 40.7, 46.9), human CA II (115456, 37.5, 45.5), human CA III (1070516, 36.2, 45.1), human CA IV (115465, 25.9, 35.7), human CA V (461680, 31.0, 37.5), human CA VB (6005723, 34.1, 41.0), human CA VI (1070519, 31.6, 40.2), human CA VII (1168744, 34.2, 42.5), human CA-RP VIII (461681, 28.2, 35.5), human CA IX (2135876, 29.2, 38.5), human CA-RP X ((Ref. 39), 20.2, 28.2), human CA-RP XI (3283386, 26.5, 33.0), human CA XII (2708639, 28.9, 37.2), human receptor-type tyrosine phosphatase β (190744, 23.7, 37.2), human receptor-type tyrosine phosphatase γ (292411, 27.3, 35.0), Klebsiella pneumoniae (2773319, 39.1, 46.9), macaque CA I (461679, 38.5, 46.9), mouse CA I (1345656, 36.3, 44.5), mouse CA II (115457, 38.1, 45.6), mouse CA III (115463, 37.3, 44.9), mouse CA IV (2493487, 27.8, 36.7), mouse CA V (1168742, 28.2, 36.7), mouse CA VI (18761, 29.8, 37.2), mouse CA VII ((Ref. 40), 37.4, 44.9), mouse CA-RP VIII (115476, 30.5, 37.4), mouse CA-RP XI (AF050105, 28.1, 34.2), mouse CA XIV (BAA78709, 29.1, 36.5), mouse receptor-type tyrosine phosphatase γ (293774, 29.2, 36.5), Neisseria gonorrhoeae (1841441, 36.4, 45.6), Pinctada fucata (1480031, 20.0, 30.0), rabbit CA I (115452, 36.8, 46.8), rabbit CA II (1168741, 39.1, 46.2), rabbit CA IV (1345657, 26.3, 33.2), rat CA II (115459, 37.2, 44.7), rat CA III (2708636, 38.2, 45.3), rat CA IV (1345658, 26.5, 33.2), rat CA V (1168743, 34.6, 41.5), rat receptor-type tyrosine phosphatase β (487781, 25.5, 34.4), rhesus CA I (115450, 38.7, 47.1), sheep CA I (1345651, 38.3, 46.1), sheep CA II (115460, 36.9, 44.0), sheep CA VI (115469, 34.8, 40.0), Sheep CA-RP XI (Y07785, 28.8, 35.0), Synechococcus PCC7942 (18254581, 37.6, 43.9), vaccinia virus (335595, 20.0, 35.0), variola virus (416745, 25.0, 41.7), and Vibrio cholerae (The Institute for Genomic Research, 35.1, 42.3).