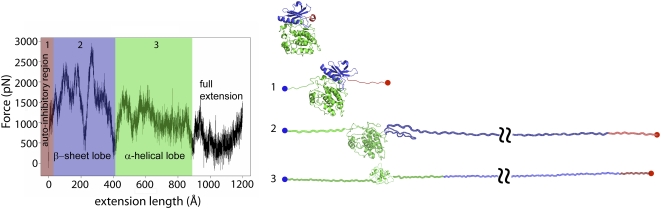
FIGURE 6.
Constant velocity steered molecular dynamics simulation of the mechanical unfolding of twitchin kinase. (Left) Force-extension curve obtained from SMD simulations by stretching the twitchin kinase domain (1KOA) between its C terminus and its N terminus at a pulling speed of 0.5 Å/ps. The total simulation time was 2.4 ns using 33,428 total atoms including 18 Na+ and 9305 water molecules. The fixed atom was Tyr-5915 and the SMD atom Arg-6261. (Right) Four snapshots of twitchin kinase stretched from its termini taken at no extension (rest), after 65 Å (1), 340 Å (2), and 639 Å (3) of extension. At rest, the kinase domain is in a closed conformation. The active site is occupied by the autoinhibitory region (red), which makes extensive contact with the catalytic site, blocking substrate binding. (1) At low forces the regulatory tail will unravel reversibly and expose the active site to its substrates. (2) At high forces the kinase begins to unfold and the integrity of the active site is disrupted. The small lobe (blue), made mainly of β-sheets, unravels first followed by the unfolding of the α-helical rich large lobe (green).