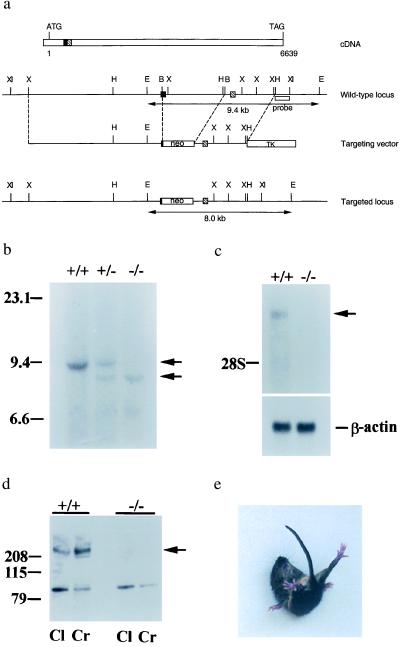
Figure 1.
Characterization of the mouse Ca2+ channel α1A null mutation. (a) The start and the stop codons are indicated in the cDNA map. The solid box and the hatched box represent the corresponding exons in the genomic DNA locus. Double arrowhead lines under the wt and the targeted locus represent the expected fragments when digested with EcoRI and hybridized with the probe (open box underneath the wt locus). B, BalI; E, EcoRI; H, HindIII; X, XbaI; XI, XhoI. (b) Southern blot analysis of offspring derived from intercrosses of calcium channel α1A heterozygous parents. The 9.4-kb fragment is the wt allele and the 9.0-kb fragment is the targeted allele. +/+, wt; +/−, heterozygote; −/−, mutant. (c) Northern blot analysis of brain tissues. One-microgram aliquots of poly(A) RNA from wt and −/− animals were probed with a 410-bp fragment from the 5′ region of rat brain α1A cDNA. The lower bands are β-actin mRNA, used as a control. (d) Western blot analysis of membrane fractions from cerebral (Cr) and cerebellar (Cl) cortices of wt and α1A−/− mice. The band observed near 100 kDa in α1A−/− is unlikely to correspond to the glycosylated, 95-kDa short form of α1A (19) because it was eliminated after purification by affinity to wheat germ-agglutinin-Sepharose. Furthermore, no reverse transcription–PCR products were obtained from α1A−/− mRNA with various primer sets targeting the first half of the channel, in contrast to the finding of PCR products of correct length when wt mRNA was tested (see Methods). (e) An α1A mutant mouse attempting to right itself during a falling episode.