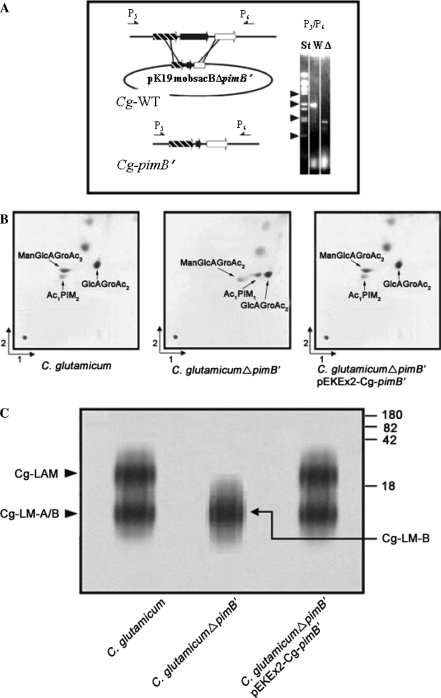
Fig. 2.
Construction of inframe deletion mutant of pimB′ in C. glutamicum and resulting phenotype. (a) Strategy to delete pimB′ by use of vector pK19mobsacBΔpimB′ by two homologous recombination events with the wild type chromosome C. glutamicum (Cg-WT). The deletion is demonstrated on the right via PCR using primer pairs P5/P6 showing the expected 1088 bp fragment for the deletion mutant in the lane marked “Δ”, and that of 2159 bp for the wild type marked “W”. The lane marked “St” is the standard consisting of BstEII-fragments of λ-DNA, with arrowheads positioned at 0.70, 1.37, 2.32, and 3.68 kb. (b) TLC-analysis of PIM biosynthesis in C. glutamicum, C. glutamicumΔpimB′ and C. glutamicumΔpimB′ pEKEx2-pimB′. Glycolipids were visualized by spraying plates with α-naphthol/sulfuric acid, followed by gentle charring of TLC plates. (c) Lipoglycan profiles of C. glutamicum strains analyzed using SDS-PAGE and visualized using a Pro-Q emerald glycoprotein stain (Invitrogen) specific for carbohydrates. The major bands represented by Cg-LAM, Cg-LM-A, and Cg-LM-B are indicated. The four major standard bands indicated on the side of the gel represent glycoproteins of 180, 82, 42 and18 kDa, respectively