Figure 8.
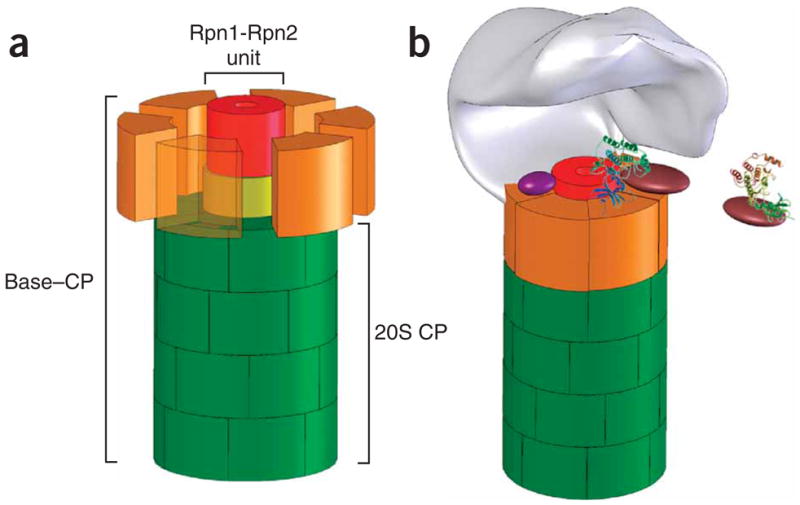
A model for how Rpn1-Rpn2 mediates substrate reception and translocation through the proteasome. (a) The central unit within the base. In a schematic structural model of the base (roughly drawn to scale), Rpn2 (yellow) attaches to the center of the 20S CP surface (green). Rpn1 (red) sits atop Rpn2. Rpt ATPases (orange; dispersed, with one transparent for clarity) attach to the 20S surface, wrapping around Rpn1-Rpn2 to complete the dome-like base. The model is a simplification based on the most structured regions of these proteins that are picked up in our assays. We do not exclude that both Rpn1 and Rpn2 may have extended flexible regions for interaction with auxiliary factors that protrude beyond the ring-like frame. (b) The 19S RP revisited. A lid subcomplex (schematically shown in gray) entombs a cavity within the 19S by attaching asymmetrically to the base15,29–31. The resulting interface between the lid and base accommodates interchanging proteasome auxiliary factors and substrate delivery proteins (for example, purple and brown), many of which are nominally part of the base. The protruding rim of the Rpn1-Rpn2 stalk may serve as a docking site for substrate-recruitment factors, relaying substrates to the ATPase domain for unfolding and translocation through the extended proteolytic channel. Both the ATPase domain and the newly identified central unit partake in translocation, although the precise trajectory is yet to be charted.
