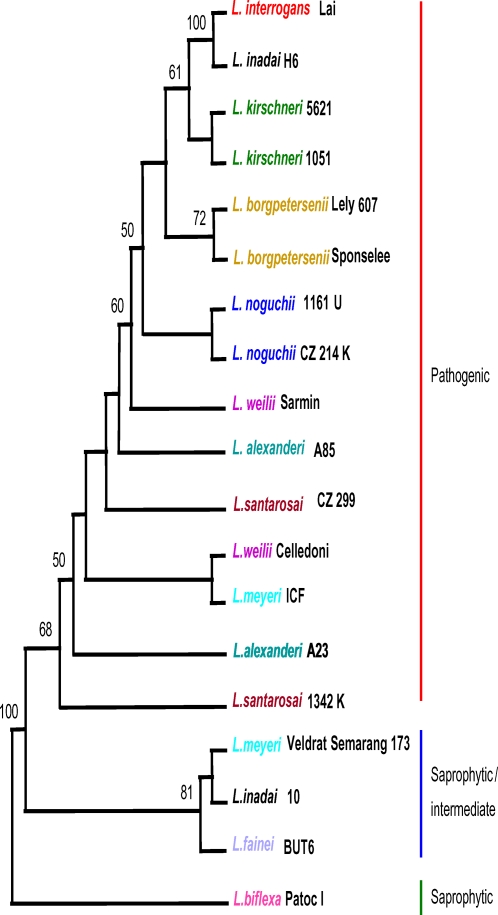
Figure 1. Consensus tree based on PCR amplification data.
Majority-rule consensus tree elaborated under the parsimony criterion and based on binary data (absence/presence) coded from amplification patterns in the S10-spc-α locus for different Leptospira species. Numbers on nodes are bootstrap support after 100 replicates. Only bootstrap values above or equal to 50% are shown. Species included in the sequence analysis are coded in color. L. biflexa was used as the outgroup. CI = 0.346.