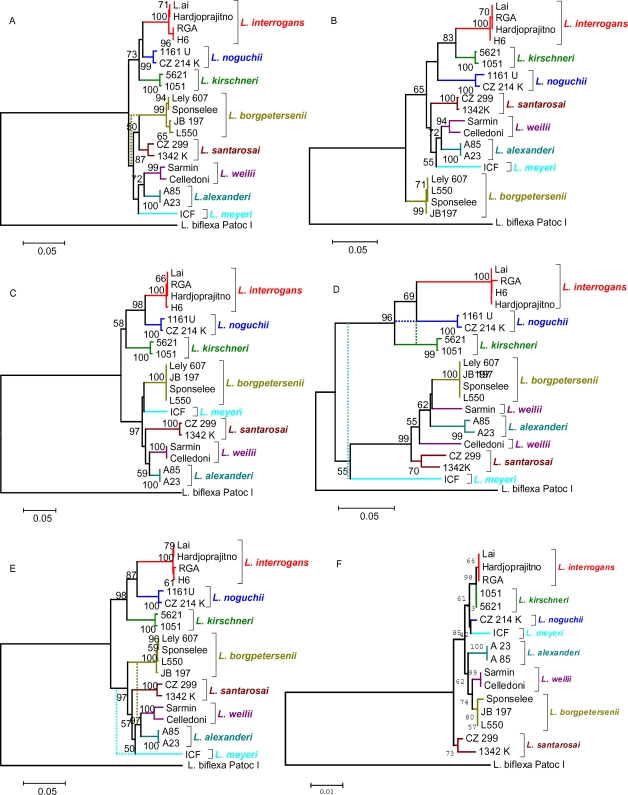
Figure 2. Phylogenetic trees based on Tamura-Nei distances and elaborated using the Neighbor-Joining method.
Distances were calculated from the 300–301 (A), 621–625 (B), 624–650 (C) and G1–G2 (D) sequence fragments within the S10-spc-α locus of pathogenic species of Leptospira. The total evidence was combined and analyzed under identical conditions (E). In addition, data available from 16S rDNA (rrs) sequences were used to obtain an alternative hypothesis for the relationships of diverse Leptospira strains (F). Dotted lines show alternative branching patterns, with bootstrapping values ≥50%, obtained in the consensus majority rule tree obtained by parsimony criterion. Numbers above branches represent the percentage of bootstrapping results (2000 replicates). Trees are drawn to scale as indicated by the bar depicted below each tree; bars represent the estimated distance in units of the number of base substitutions per site. The scale the 16S rRNA-based tree is expanded relative to other loci. L. biflexa was used as the outgroup.