Abstract
Maternal-Effect Dominant Embryonic Arrest (“Medea”) factors are selfish nuclear elements that combine maternal-lethal and zygotic-rescue activities to gain a postzygotic survival advantage. We show that Medea1 activity in Tribolium castaneum is associated with a composite Tc1 transposon inserted just downstream of the neurotransmitter reuptake symporter bloated tubules (blot), whose Drosophila ortholog has both maternal and zygotic functions. The 21.5-kb insertion contains defective copies of elongation initiation factor-3, ATP synthase subunit C, and an RNaseD-related gene, as well as a potentially intact copy of a prokaryotic DUF1703 gene. Sequence comparisons suggest that the current distribution of Medea1 reflects global emanation after a single transpositional event in recent evolutionary time. The Medea system in Tribolium represents an unusual type of intragenomic conflict and could provide a useful vehicle for driving desirable genes into populations.
Keywords: postzygotic, selfish gene, Tribolium, gene driver
Propagation of selfish elements nonessential for host survival can be mediated either cytoplasmically or chromosomally. Many cytoplasmically inherited Wolbachia or microsporidia enhance their own propagation by feminizing, sterilizing, or killing infected males or male gametes, thereby redirecting investment toward production of (infected) females and ensuring a more rapid spread of the pathogen than could be achieved solely by passive transmission in the cytoplasm (1). In other cases such a transmission advantage is achieved by sperm from Wolbachia-infected males conferring inviability on eggs laid by uninfected mates (2). Chromosomally inherited Segregation Distorter (SD) and sex-ratio meiotic drive alleles in Drosophila create a prezygotic transmission advantage in heterozygous males by conferring inviability on rival spermatids bearing the alternative alleles (3–5).
We discovered the existence of a novel category of selfish genetic elements that act postzygotically and include a maternal and a zygotic component, both of which are required for expression of selfish behavior (6, 7). These factors, which we termed Medea (M) elements, are widespread in natural populations of Tribolium flour beetles but are otherwise unknown in the invertebrate world (8). Our initial discovery (6) was followed by two reports describing similar phenomena in mice. The first involved a spontaneous chromosome 8 mutation that occurred only once in a single laboratory strain of Mus musculus and is associated with a maternally conferred autoimmune disease, severe combined anemia and thrombocytopenia (scat) (9). A second example, also from M. musculus, involves the homogeneously staining region (HSR) on chromosome 1, which consists of variable numbers of tandem copies of a 100-kb repeat. Like Medea and scat, some variants of HSR impart maternal lethality to late embryos, which is prevented by a zygotic copy of the HSR chromosome inherited from either parent (10–12). The mechanisms of postzygotic self-selection in these systems are still unknown.
Because no molecular mechanism has yet been reported for any maternal selfish gene, we decided to identify the molecular lesion associated with the Medea factor M1 on the third linkage group of Tribolium castaneum. Our findings indicate that insertion of a very large, composite Tc1/mariner transposon is tightly associated with M1 activity and is the probable cause of the maternally controlled selfish behavior of this locus. This insertion appears to have occurred only once, followed by worldwide spread of the affected chromosome.
Results
Positional Cloning of Medea1.
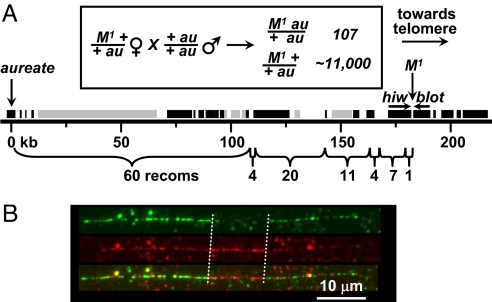
We positionally cloned a region encompassing the site of the M1 locus on the third linkage group of T. castaneum by performing a chromosome walk in a non-M BAC library. The walk spanned a total of >700 kb and was oriented and monitored by high-resolution recombinational mapping using “left”-flanking (centromere-proximal) markers only, because no right-flanking, visible mutant markers were available. This mapping yielded 107 recombination events in the 182-kb segment bounded by the recessive visible aureate mutation and the estimated position of the target M1 locus (Fig. 1A). Thus, the mean distance between recombination events was 1.7 kb. This process confined the probable site of the M1 trait to a small region flanked by the 3′ ends of the Tribolium orthologs of the Drosophila genes highwire (hiw) (13) and bloated tubules (blot) (14), closely juxtaposed in tail-to-tail orientation (Fig. 1A).
Fig. 1.
M1 is associated with an insertion. (A) aureate-to-Medea1 region of the chromosome walk in the non-M1 GA2 strain. Brackets below the chromosome show the distribution of 107 aureate (au)-Medea1 (M1) recombination events among seven segments constituting the 182-kb region. The recombinants were found among ≈11,000 progeny of the indicated backcross (boxed) of the transheterozygote to the double recessive. Bars above the chromosome indicate transcribed regions predicted by GLEAN (25) with support from ESTs, cDNAs, or tiling array data (black bars) or without such support (gray bars). The position of the M1 locus just downstream of the blot gene in the non-M1 strain was predicted from the distribution and average spacing (1.7 kb) of recombination events. Directions of transcription of hiw and blot are indicated by horizontal arrows. The site of the au mutation is set at “zero” on the kb scale. (B) Cytological visualization of the M1 lesion. The panel depicts a single M1 chromosome fiber (24). Fibers were hybridized to a non-Medea (GA2-strain) BAC clone (green fluorescence, g) and an M1 BAC clone (red fluorescence, r). Both BAC clones include the M1 insertion point and >30 kb of flanking sequence on each side. The digital overlay (o) of the red and green images clearly indicates the presence of a large insertion in the M1 fiber (delimited by vertical dashed lines), which does not hybridize with the GA2-derived probe. For the red-filtered image, note the relative enhancement of red fluorescence in the insertion region, in which there is no competition for green-labeled probe.
M1 Colocalizes with a Large Insertion.
The corresponding region of an M1 strain was obtained by screening an M1 BAC library with a hiw-specific probe. A 129-kb BAC clone was identified and confirmed to contain the region of interest by hiw- and blot-specific PCR, as well as PCR specific for regions upstream of blot. The clone was completely sequenced and provided ≈40 kb of M1 sequence upstream (telomeric) of blot as well as ≈60 kb upstream (centromeric) of hiw. Inspection of the M1 sequence revealed one striking difference between M1 and non-M1 strains, namely the presence in the M1 strain of a 21.5-kb insertion of exogenous sequence at precisely the calculated position of the M1 locus as determined by high-resolution genetic mapping. To verify the presence of the M1 insertion by direct visualization, we performed fluorescence in situ hybridization to M1 chromosome fibers (fiber-FISH). The presence of a large insertion of exogenous DNA was indeed confirmed (Fig. 1B).
The M1 Insertion Is a Composite Tc1 Transposon.
Sequence analysis indicated that the entire 21.5-kb insertion comprises a large, composite Tc1 transposon, i.e., a transposon within a transposon (Fig. 2). The hiw-to-blot intergenic region of the non-M strain GA2 is depicted in Fig. 2A. The region spans 776 nt flanked by the 3′ UTRs of hiw and blot. The insertion point of the 21.5-kb segment is approximately midway between the hiw and blot polyadenylation signals. The centromere-proximal end of the insertion consists of a full-length (1.4-kb) Tc1 element encoding a nearly intact, 346-residue transposase [Fig. 2B and supporting information (SI) Figs. S1 and S2]. A single-base transition, C→T, which converts R to a stop codon at residue 182, is the only apparent defect. The 50 nt at the telomere-proximal end of the 21.5-kb insertion consists of an intact, Tc1 right terminus including a full, 35-nt inverted terminal repeat (ITR). DNA transposons of the Tc1/mariner/IS630 superfamily are the most plentiful found in the Tribolium genome, being distributed among 30 subfamilies (15).
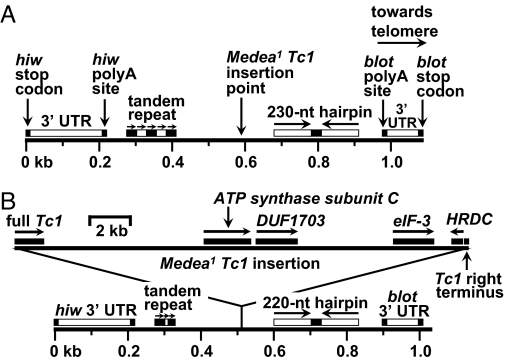
Fig. 2.
The M1-associated insertion is a 21.5-kb composite Tc1 element. (A) Map of hiw-to-blot region of a wild-type chromosome. The two genes (tail-to-tail orientation) are separated by a span of 1.1 kb (stop codon to stop codon). Vertical arrows indicate stop codons, polyA addition sites, and Tc1 insertion site. The insertion point is flanked by five copies of a 27-nt, imperfect tandem direct repeat, as well as a 230-nt hairpin consisting of a 20-nt central loop flanked by a pair of 105-nt, imperfectly palindromic stems. Horizontal arrows indicate the tandem direct repeats, hairpin stems, and orientation of the entire segment on the chromosome. Horizontal bars above insertion target chromosome denote main features of region flanking the M1 insertion target. (B) Map of hiw-to-blot region of an M1 chromosome. Solid bars above the 21.5-kb composite Tc1 insertion (top part of B) denote genes or gene fragments within the insertion. Horizontal arrows indicate directions of transcription. Of these genes (Tc1 transposase, ATP synthase subunit C, DUF1703, eIF-3, and HRDC) only DUF1703 is thought to be potentially functional (see Fig. S3). The isolated Tc1 right terminus is not shown to scale. Note that the tandem repeat and hairpin are shorter in the M1 strain than in wild type (compare A and B). Also note the different scales for insertion vs. flanking regions.
M1-Associated Tc1 Transposon Family Members Are Nondegenerate.
A total of five closely related copies of the M1-associated Tc1 element are present in the GA2 genome assembly. All five are full-length, and two of the five have intact coding sequences and potentially encode functional transposase (Fig. S2). The five are members of the Tc1-DD34E subclass (16). In view of the dearth of accumulated mutational defects in members of this subclass, it is likely that this element invaded the Tribolium genome in recent evolutionary time, quite possibly since the speciation of castaneum. We could find no examples of highly degenerate members of this Tc1 subclass or of any large, composite Tc1 transposons in the GA2 genome.
The M1 Insertion Contains Defective Copies of Vital Genes.
We determined that no well conserved copy of the 20-kb central segment (forming the “cargo” of the composite Tc1 element) exists in the non-Medea (GA2) genome but that there are regions of extensive similarity on three other linkage groups (LG), namely 6, 8, and 10. This central segment contains incomplete or defective copies of at least three genes, namely ATP synthase subunit C (ATPsyntC), elongation initiation factor 3 (eIF3), and a gene encoding a helicase RNaseD C-terminal domain (HRDC) (Fig. 2B). Orthologs of ATPsyntC and eIF3 are vital in Drosophila, and intact copies occur elsewhere in the GA2 genome (Fig. S3).
The M1 Insertion Contains a Bacterial Gene.
In addition to the defective ATPsyntC, eIF3, and HRDC-motif genes, there is a fourth gene, previously known only from bacteria. Sequence analysis indicates that this is the only potentially functional gene present in the central cargo segment of the composite Tc1 insertion. The protein encoded by this intronless gene shows no similarity to any known or predicted animal protein but is related to a group of proteins containing the bacterial domain of unknown function DUF1703 (pfam08011) (Fig. S4). DUF1703 proteins have recently been proposed to be members of the PD-(D/E)XK nuclease superfamily (17). Before our analysis, DUF1703 genes were known only in prokaryotes (http://pfam.sanger.ac.uk/family?entry=pf08011&type=Family), the sole exception being a single, highly truncated fragment found in the bacterivorous amoebozoan Hartmannella vermiformis (18). The (non-Medea) Tribolium genome assembly contains four apparently intact copies of the DUF1703 gene that are closely related to the M1 copy, showing ≈90% sequence identity over almost 700 amino acid residues. At least two of these are transcriptionally active, both on LG6. There are also four truncated copies that are largely identical to corresponding regions of the M1 copy and ≈10 additional copies more distantly related (20–30% identical at the amino acid level). These more distantly related copies are clearly also members of the bacterial DUF1703 family (Fig. S4). We are unaware of other examples of lateral transfer of a prokaryotic gene into an animal genome followed by expansion in copy number of the invading gene. The grouping of the Tribolium DUF1703 proteins into a distinct clade suggests either a recent gene family expansion in the beetle's genome or evolutionary constraints imposed on a functionally related group.
The M1 Insertion Occurred Once, Then Spread Globally.
Sequence comparisons among geographically diverse strains of T. castaneum indicate a single origin of M1 followed by global radiation. We compared sequences flanking the site of the M1-associated Tc1 insertion from 23 strains of T. castaneum collected in 15 countries worldwide and previously categorized as M1 (11 strains) or non-M1 (12 strains) based on an in vivo genetic test for maternal lethality (see Materials and Methods). Sequence analysis confirmed earlier diagnoses of Medea genotypes (8). Fig. 3 shows the results of an unrooted cluster analysis of sequence relatedness in the region of the Tc1 insertion point. A striking observation that emerges from this comparison is that M1 strains are all nearly identical in sequence and constitute a single, distinct clade despite their widespread global distribution, spanning Central America, South America, Africa, East Asia, and Southeast Asia. Non-M1 strains of equally diverse origins constitute a second, more highly variable clade. This suggests a single origin of the M1 insertion followed by worldwide spread in recent evolutionary time, as has been suggested for organophosphate insecticide resistance in Culex pipiens (19). Sequence variation among non-M1 strains, as well as differences between M1 and non-M1, rapidly diminish >300 nt from the insertion point on either side (Fig. S5). The clade differentiation between M1 and non-M1 is not apparent in the 3′ UTRs of the flanking blot and hiw genes.
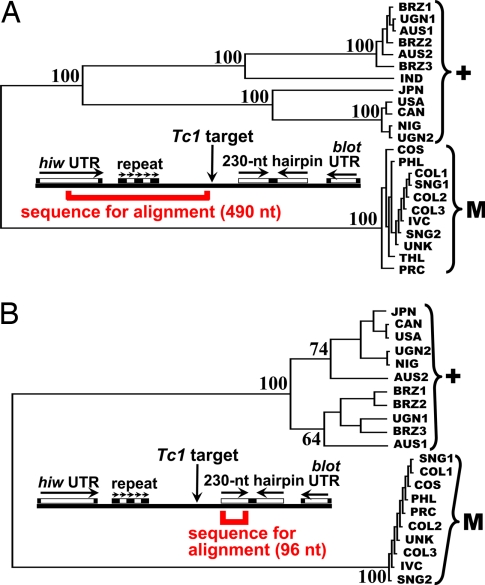
Fig. 3.
Phylogenetic analysis of insertion-site sequences. Shown are unrooted phylogenetic trees based on multiple alignments of sequences flanking both sides of the Tc1 insertion point. Trees are shown in Phylip format, with bootstrap values indicated. Data in A are based on “left”-flanking sequences (centromeric side of insertion), and data in B are based on right-flanking sequences (telomeric side of insertion). Segment lengths refer to the non-Medea GA2 (USA) strain. Countries of origin include Brazil, Uganda, Australia, India, Japan, the United States, Canada, Nigeria, Costa Rica, the Philippines, Colombia, Ivory Coast, Singapore, Thailand, and Peoples' Republic of China, abbreviated BRZ, UGN, AUS, IND, JPN, USA, CAN, NIG, COS, PHL, COL, IVC, SNG, THL, and PRC, respectively. UNK was collected by Alexander Sokoloff before 1970 and is of unknown origin. The USA (GA2) strain, which was the source of the whole-genome sequence, was collected in 1983. All other strains were collected in 1988. Brackets to the right indicate grouping of M1 (“M”) and non-M1 (“+”) strains, as determined by genetic testing (see Materials and Methods).
M1 Is Sandwiched Between Two Genes with Both Maternal and Zygotic Components.
The blot gene, like Medea itself, has both maternal and zygotic functions, at least in Drosophila (14). The Drosophila Highwire protein negatively regulates synaptic proliferation at neuromuscular junctions by binding to the Smad protein Medea (Med, no apparent relation to Medea in Tribolium despite the coincidence of names). In Drosophila, Med mediates synaptic proliferation via the BMP signaling cascade. Although Drosophila highwire is apparently a strictly zygotic gene, its binding partner Med is required both maternally and zygotically (20). However, there is no obvious mechanism that connects the mixed maternal/zygotic attributes of either blot or hiw with those of the closely juxtaposed Medea locus.
Discussion
Chen et al. (21) constructed a synthetic Medea factor in Drosophila based on our conception of a maternal poison acting in concert with a tightly linked zygotic antidote. The lethal component in this design was a maternally expressed microRNA (miRNA) that silenced the maternally required myd88 gene. The rescue component was a zygotically expressed variant of myd88 that carried a deletion rendering it insensitive to the miRNA toxin. The (early) zygotic expression of the myd88 antidote proved effective in fulfilling a function that is normally provided maternally. As predicted (7), this artificial Medea chromosome supplanted its nonselfish homolog in population cage studies. It is unknown whether Medea in Tribolium functions in a similar manner or by some still-unforeseen mechanism. Although there are several pseudogenes and one potentially intact gene within the M1 insertion, the transcriptional activity of these regions has not been examined.
One way to gain insight into the Medea mechanism might be through molecular analysis of Medea revertants. We have isolated five such mutations after γ-irradiation of M1 males. Each revertant chromosome has lost maternal-lethality while retaining zygotic rescue (unpublished observations). Identification of the underlying molecular lesions and sequence analysis of the insertion regions of revertant chromosomes could reveal motifs required for M-related lethality.
In addition to M1 there is at least one other Medea factor, namely M4, that could be targeted for positional cloning. The sequence of the M4 locus is unknown, but it is the most prevalent Medea factor found in natural populations of Tribolium worldwide (8). Like M1, it maps near one extreme end of the third linkage group, but on the opposite chromosome arm. These two factors display identical maternal-lethal and zygotic-rescue behavior but do not cross-rescue: a zygotic copy of M1 does not prevent the maternal-lethal action of M4, and vice versa (8). Interestingly, although they function independently and can easily be isolated from double-Medea strains by genetic segregation, M1 has never been found in nature in the absence of M4 (8).
Our data provide very strong evidence for a single origin of M1 followed by global dispersal. The current, patchy distribution of M1 (and perhaps also M4) might exist as a remnant of the original patterns of global spread, or it could reflect regional differences in the outcome of conflict between Medea and non-Medea genotypes. One evolutionary hypothesis is that each Medea element arose as a single mutational (insertion) event within a population and then spread. An alternative hypothesis is that the Medea type sequence was fixed in a species or population where it had no effect, but then a hybridization event occurred, perhaps even across species, and in the new genetic background it expressed its selfish behavior. In this scenario, one can imagine that the Medea type species carried a version of chromosome 3 that was fixed for both M1 and M4 and that the current distribution of M strains, in which M1 is confined to a subset of the regions where M4 is fixed, derived from secondary loss of M1 in some areas after the selfish M1 M4 chromosome had spread widely.
A complete understanding of the Medea syndrome will not be attained without additional studies of the hybrid incompatibility factor “H” on the ninth linkage group (22). This factor, naturally occurring in several Tribolium strains from India where Medea is notably absent, is so named because of its incompatibility with either of the third linkage group Medea factors M1 or M4 (23). Hybrid beetles carrying one copy of H and one copy of either M1 or M4 die as larvae. In the presence of the H factor, the lethal effect of M is zygotic rather than maternal, and there is no zygotic protection. The discovery of H-incompatibility, the positional cloning and molecular characterization of the M1 locus, and the isolation of several M1 knockout mutations coupled with the completion of the genome sequence, the availability of expression microarrays, the development of efficient germ-line transformation protocols, and the power of RNA interference in Tribolium at long last provide the tools required to examine the underlying mechanisms for this unique and fascinating phenomenon.
Materials and Methods
BAC Library Construction and Positional Cloning.
A genomic BAC library was prepared from the highly inbred, non-M strain GA2. High-molecular-weight DNA was partially digested by using a combination of EcoRI and EcoRI methylase, then size-fractionated by pulsed-field gel electrophoresis. DNA fragments were cloned into the pBACe3.6 vector between the two EcoRI sites. A sequence tagged site ≈1 cm from the target was used to initiate the bidirectional walk. For each step the library was screened with overgo probes derived from BAC end sequences. The BACs were ordered and oriented by PCR of end sequences, and the walk was oriented by analysis of recombination events occurring within the BAC contig (Fig. 1A). After identifying the correct orientation, the walk was continued unidirectionally. The walk hit an obstruction in the telomere-proximal direction (toward the target M1 locus) in the form of a highly repetitive region that could not be crossed in the EcoRI library. This obstacle was circumvented by taking one step in a HindIII library, after which the walk resumed in the EcoRI library, proceeding to a point beyond the target M1 locus. A second genomic BAC library was prepared by Amplicon Express, from the Singaporean M1 strain in the pECBAC1 cloning vector. Genomic DNA was partially digested with MboI and ligated into the BamHI cloning site.
Sequence Analysis.
The M1 BAC was sequenced and assembled by the Human Genome Sequencing Center at Baylor College of Medicine. The 21.5-kb insertion was compared with the sequenced T. castaneum genome (BLASTN), and the insertion sequence, as well as the corresponding GA2 regions, were analyzed for the presence of known genes, gene predictions, or other potential coding regions by tBLASTN and BLASTX. DNA templates obtained from world strains were sequenced by using an ABI 373A DNA sequencer (College of Veterinary Medicine, Kansas State University).
Fiber Fluorescence in Situ Hybridization (Fiber-FISH).
Chromosome fibers were prepared from M1-strain, whole fifth-instar larval nuclei using the method of Jackson et al. (24). Fibers were hybridized to the biotin-conjugated (green fluorescent) GA2-strain (non-Medea) BAC clone 40D19 (GenBank accession no. AC154135), which includes the empty site of the M1-associated insertion, and digoxigenin-conjugated (red fluorescent) M1-strain BAC clone 22J23 (GenBank accession no. AC205539), which contains the insertion.
Phylogenetic (Cluster) Analysis of Insertion-Site Sequences.
Multiple sequence alignments (AlignX; Invitrogen, followed by manual adjustment) were used to construct unrooted phylogenetic trees using TreeTop (www.genebee.msu.su/services/phtree_reduced.html) with the following settings (extra tree format: Phylip; picture format: Unrooted). Separate alignments were made based on the 490-nt segment extending from the middle of the hiw 3′ UTR to the Tc1 insertion point (centromeric side of insertion) and on the 96-nt segment that comprises the left stem of the 230-nt hairpin (telomeric side of insertion). Sequences were derived from 12 non-M1 and 11 M1 strains collected in 15 countries.
Correlation Between Tc1 Insertion and Medea Genotype in Wild-Caught T. castaneum.
Twenty-three strains from 15 countries worldwide were tested for Medea1-associated maternal-lethal activity, then subjected to sequence analysis in the vicinity of the insertion site. Maternal-lethal activity was tested by “marker exclusion” as follows: Crosses were established in triplicate between females from each test strain and non-M1 3P2/au14 males (one female and two males for each cross), where 3P2 represents a dominantly marked, second linkage group balancer tightly linked to M1, and au14 is a lethal balanced by 3P2. Females were allowed to mate and oviposit for 10 days, after which time genomic DNA was isolated from each female for PCR-sequence analysis (see below). F1 female 3P2/* progeny were collected from each cross, where * indicates the test chromosome (M1 or non-M1). The females were test-crossed in small groups to non-M1, mas p au males (five of each sex). At least 100 progeny from each test cross were collected and scored for the dominant 3P2 phenotype. Non-Medea lines were expected to produce ≈50% 3P2-bearing progeny, whereas Medea1 lines were expected to produce no 3P2 progeny, because of M1-induced maternal lethality to zygotes bearing this non-Medea chromosome.
After the above-described genetic diagnoses, females were tested for the presence of the Tc1 insertion by PCR, and sequences flanking the insertion sites or insertion junctions were determined. PCR was done in two ways: in the first experiment, the PCR mixture had three primers, namely a forward hiw primer common to M1 and non-M1, a reverse primer specific for M1, and another reverse primer specific for non-M1. In this way heterozygotes could be differentiated from either homozygote in a single reaction. In addition, we performed separate PCRs specific for M1 and non-M1. All experiments were double-blind. See the Fig. 3 legend for strain names and geographic origins.
Supplementary Material
Acknowledgments.
We thank Sue Haas, Sara Brown, Beth Stone-Smith, Kelli Goodrich, and Barb van Slyke for technical support and Dr. Susan J. Brown (Kansas State University) for providing the HindIII BAC library. This research was supported in part by the Exelixis Pharmaceutical Co. and by the Human Genome Sequencing Center (Baylor College of Medicine).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU597633–EU597678). GLEAN sequences are available at the Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800444105/DCSupplemental.
References
- 1.Burt A, Trivers R. Genes in Conflict. Cambridge, MA: Harvard Univ Press; 2006. pp. 179–180. [Google Scholar]
- 2.Hoffman AA, Turelli M, Harshman LG. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics. 1990;126:933–948. doi: 10.1093/genetics/126.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartl DL, Hiraizumi Y, Crow JF. Evidence for sperm dysfunction as the mechanism of segregation distortion in Drosophila melanogaster. Proc Natl Acad Sci USA. 1967;58:2240–2245. doi: 10.1073/pnas.58.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. PLoS Biol. 2007;5:2560–2575. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biol. 2007;5:2576–2588. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beeman RW, Friesen KS, Denell RE. Maternal-effect, selfish genes in flour beetles. Science. 1992;256:89–92. doi: 10.1126/science.1566060. [DOI] [PubMed] [Google Scholar]
- 7.Wade MJ, Beeman RW. The population dynamics of maternal-effect selfish genes. Genetics. 1994;138:1309–1314. doi: 10.1093/genetics/138.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beeman RW, Friesen KS. Properties and natural occurrence of maternal-effect selfish genes (‘Medea’ factors) in the Red Flour Beetle, Tribolium castaneum. Heredity. 1999;82:529–534. doi: 10.1038/sj.hdy.6885150. [DOI] [PubMed] [Google Scholar]
- 9.Peters LL, Barker JE. Novel inheritance of the murine severe combined anemia and thrombocytopenia (scat) phenotype. Cell. 1993;74:135–142. doi: 10.1016/0092-8674(93)90301-6. [DOI] [PubMed] [Google Scholar]
- 10.Weichenhan D, Traut W, Kunze B, Winking H. Distortion of Mendelian recovery ratio for a mouse HSR is caused by maternal and zygotic effects. Genet Res. 1996;68:125–129. doi: 10.1017/s0016672300034017. [DOI] [PubMed] [Google Scholar]
- 11.Winking H, et al. Polymorphic HSRs in chromosome 1 of the two semispecies Mus musculus musculus and M. m. domesticus have a common origin in an ancestral population. Chromosoma. 1991;100:147–151. doi: 10.1007/BF00337242. [DOI] [PubMed] [Google Scholar]
- 12.Weichenhan D, Kunze B, Traut W. Restoration of the Mendelian transmission ratio by a deletion in the mouse chromosome 1 HSR. Genet Res. 1998;71:119–125. doi: 10.1017/s0016672398003206. [DOI] [PubMed] [Google Scholar]
- 13.Wan HI, et al. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 14.Johnson K, Knust E, Skaer H. bloated tubules (blot) encodes a Drosophila member of the neurotransmitter transporter family required for organisation of the apical cytocortex. Dev Biol. 1999;212:440–454. doi: 10.1006/dbio.1999.9351. [DOI] [PubMed] [Google Scholar]
- 15.Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 16.Shao H, Tu Z. Expanding the diversity of the IS630-Tc1-mariner superfamily: Discovery of a unique DD37E transposon and reclassification of the DD37D and DD39D transposons. Genetics. 2001;159:1103–1115. doi: 10.1093/genetics/159.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knizewski L, Kinch LN, Grishin NV, Rychlewski L. Realm of PD-(D/E)XK nuclease superfamily revisited: Detection of novel families with modified transitive meta profile searches. BMC Struct Biol. 2007;7:40. doi: 10.1186/1472-6807-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins RF, Gray MW. The frequency of eubacterium-to-eukaryote lateral gene transfers shows significant cross-taxa variation within amoebozoa. J Mol Evol. 2006;63:801–814. doi: 10.1007/s00239-006-0031-0. [DOI] [PubMed] [Google Scholar]
- 19.Raymond M, Callaghan A, Fort P, Pasteur N. Worldwide migration of amplified insecticide resistance genes in mosquitoes. Nature. 1991;350:151–153. doi: 10.1038/350151a0. [DOI] [PubMed] [Google Scholar]
- 20.Das P, et al. The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development. 1998;125:1519–1528. doi: 10.1242/dev.125.8.1519. [DOI] [PubMed] [Google Scholar]
- 21.Chen CH, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 22.Thomson MS, Friesen KS, Denell RE, Beeman RW. A hybrid incompatibility factor in Tribolium castaneum. J Hered. 1995;86:6–11. [Google Scholar]
- 23.Thomson MS, Beeman RW. Assisted suicide of a selfish gene. J Hered. 1999;90:191–194. doi: 10.1093/jhered/90.1.191. [DOI] [PubMed] [Google Scholar]
- 24.Jackson SA, Wang ML, Goodman HM, Jiang J. Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome. 1998;41:566–572. [PubMed] [Google Scholar]
- 25.Elsik CG, et al. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.