Figure 4.
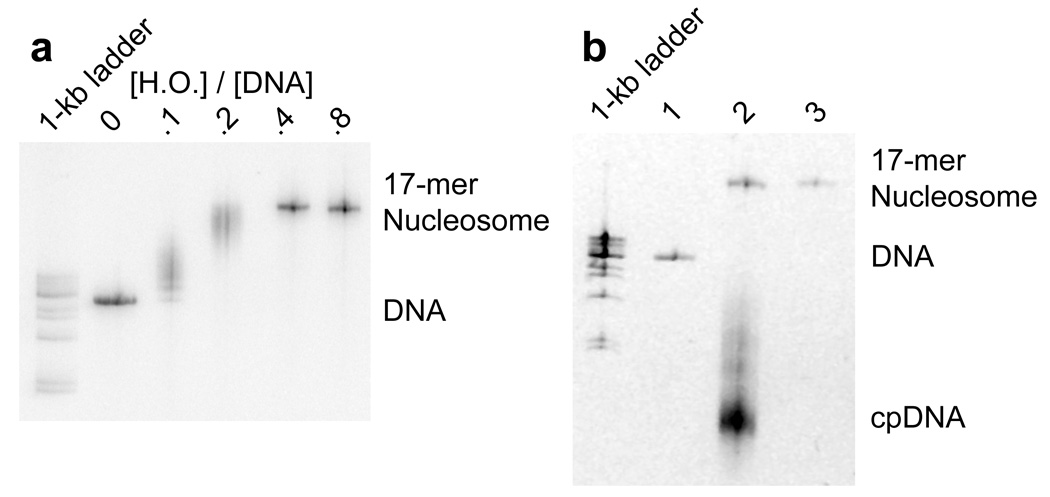

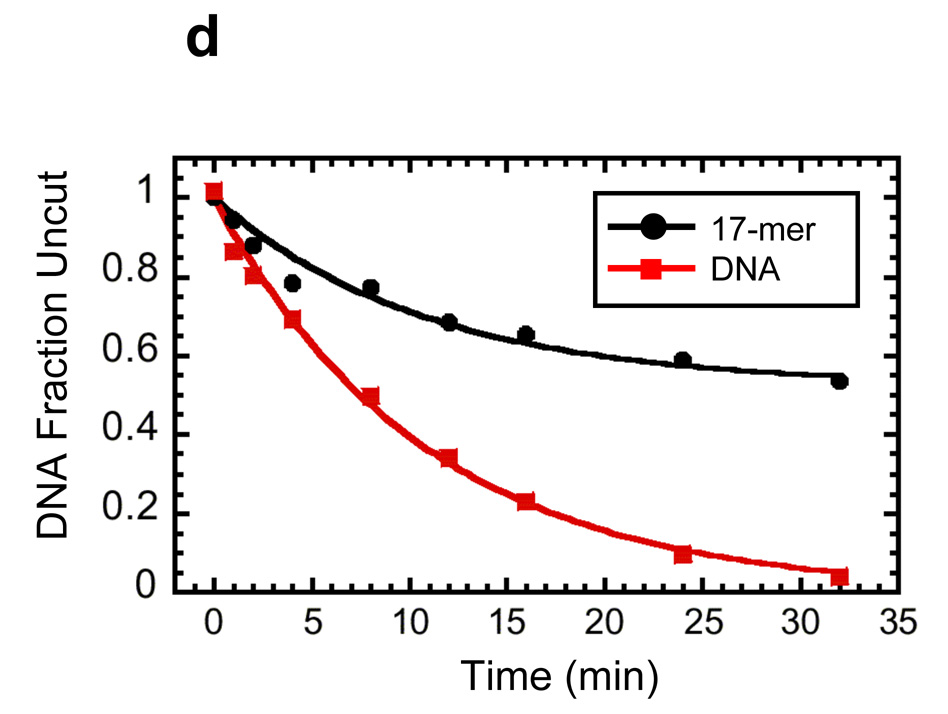
Reconstitution, purification, and characterization of nucleosome 17-mers. (a) Native gel analysis of 17-mer template DNA reconstituted with increasing concentrations of histone octamer. The mass ratio (w/w) of histone octamer to total DNA used in the reconstitution reaction in each lane is indicated. The decrease in mobility is due to nucleosome formation along the DNA template. The mass ratios 0.1 and 0.2 have wide range of gel mobilities owing to the inherent heterogeneity of DNA templates that are partially filled with nucleosomes. In contrast, the mass ratios of 0.4 and 0.8 yield a well-defined shift in gel mobility, suggesting that the DNA template is saturated with nucleosomes. (b) Ethidium-stained native gel analysis of the 17-mer DNA template (lane 1), and reconstituted nucleosome array (0.8 histone to DNA mass ratio) before (lane 2) and after purification (lane 3). The sucrose gradient removes the short low affinity competitor DNA and any aggregates. (c) Native 5% polyacrylamide gel analysis of DNA products from a Mlu I digestion of purified 17-mer nucleosomes mixed with naked DNA. The 17-mer contains 16 Mlu I sites and the naked DNA contains a single MluI site. (d) Quantification of results from (c) showing the fraction of naked DNA and the nucleosome 17-mer remaining uncut, as a function of time. The curves show best fits to the appropriate exponential decays (see Methods). These results show that 0.03 of the positioning sequences are not incorporated into nucleosomes, i.e., that 97 % of all positioning sequences in these nucleosome 17-mers are wrapped into nucleosomes.
