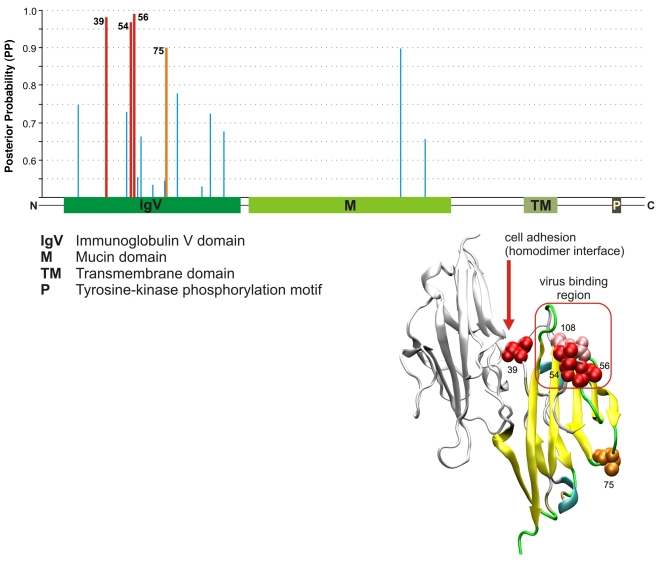
Figure 3. Structural analysis of the HAVCR1 gene.
At top is a graph showing the domain structure of the gene and corresponding Bayes Empirical Bayes [83] posterior probabilities (PP) of positive selection, based on our six-species alignments, with sites predicted to be under positive selection (PP>0.95) in red. At bottom right is a structural diagram (based on the structure of the IgV domain of the mouse gene) showing the interaction between two receptors that have been implicated in the regulation of HAVCR1's immune function. It is thought that clustering of receptors within the same cell surface might facilitate phosphorylation of the cytoplasmic tail, and that interaction between receptors from different cells might be a mechanism for B–T cell adhesion [91]. Predicted residue 39 falls within the region of these receptors, very near residue 37, which directly interacts with the opposite receptor (according to the available mouse structure). In addition, predicted residues 54 and 56 are adjacent to the virus-binding surface (shown in pink), as defined by a polymorphism in macaque [91]. Interestingly, the residue that falls between them (55) appears to be critical for virus-binding at the homologous loop in the CEA coronavirus receptor [91]. Residue 75 in the IgV domain also shows evidence of positive selection (PP>0.90, shown in orange) but its function is unknown.