Figure 5.
Molecular Analyses of Different Mutant Lines.
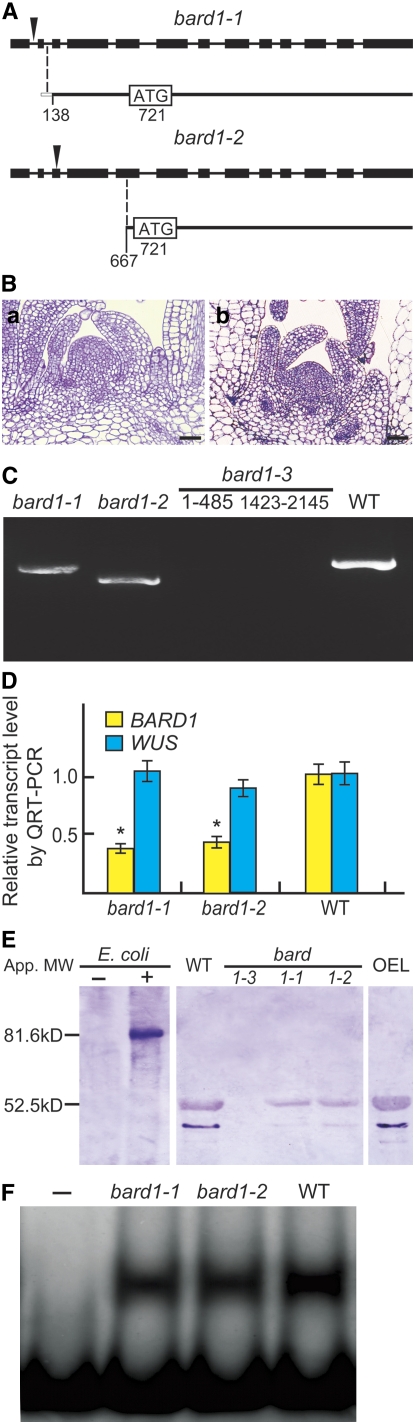
(A) Analysis of bard1-1 and bard1-2 transcripts. The 5′ RACE experiments were performed on RNA templates derived from bard1-1 or bard1-2. Inverted triangles indicate the sites of the T-DNA insertions, black boxes denote exons, and the thin lines denote introns.
(B) Median longitudinal semithin sections stained with toluidine blue showing the SAMs of 3-week-old bard1-1 (left) and bard1-2 (right). Bars = 20 μm.
(C) RT-PCR analysis of BARD1 RNAs. Full-length RNA from wild-type and truncated RNAs from bard1-1 and bard1-2 are shown. Two primer pairs derived from 5′ (1 to 485 nucleotides) and 3′ (1423 to 2145 nucleotides) regions were used to amplify bard1-3.
(D) Comparison of WUS and BARD1 mRNA levels in wild-type, bard1-1, 1-2, and 1-3 plants by QRT-PCR. Relative transcript levels (mean ± se) were calculated from triplicate QRT-PCR reactions of independent RNA samples prepared from different batches of 3-week-old Arabidopsis plants. Both WUS and BARD1 mRNA levels in the wild type were arbitrarily set to 1. *, P < 0.05 compared with the wild type.
(E) Protein gel blotting showing the presence or absence of anti-BARD1-reactive proteins in various Arabidopsis lines, as well an E. coli strain expressing a full-length ORF of BARD1. Lanes were loaded with total protein (20 μg) extracted from 3-week-old Arabidopsis lines and 10 μg total E. coli extract before (–) and after (+) IPTG induction. OEL, a protein sample was prepared from BARD1 overexpressing line (CaMV35S∷BARD1).
(F) Nuclear extracts prepared from bard1-1, bard1-2 and wild-type plants retained similar ability to form a protein-DNA complex with F4 from the WUS promoter. Nuclear extract (10 μg) obtained from 2-week-old bard1-1, bard1-2, or wild-type plants was incubated with the DNA fragment and subjected to nondenaturing gel electrophoresis as described in Methods. –, no nuclear extract was added.