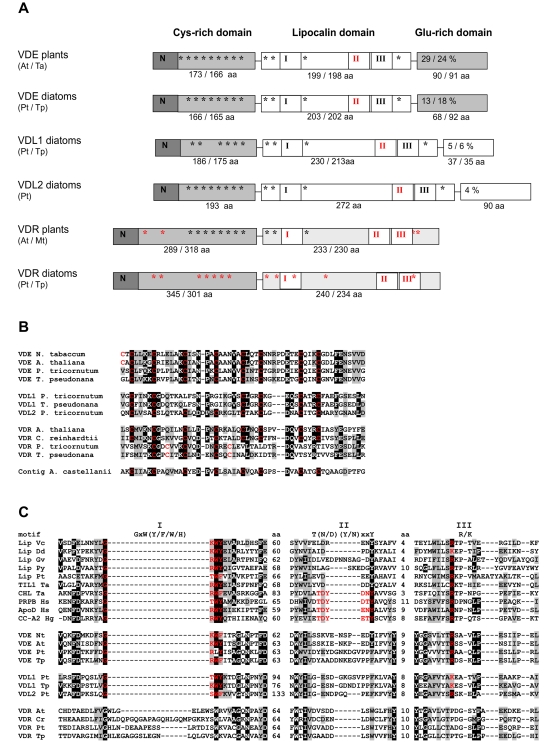
Figure 2. Domain structure of violaxanthin de-epoxidases and related proteins.
A) Schematic representation of VDE, VDL and VDR proteins (not to scale). Three different domains are shown; the cysteine-rich domains include the N-terminal targeting sequence. Black and red asterisks indicate the positions of conserved and alternative cysteine residues, respectively. The central lipocalin domain contains the lipocalin binding fold. Conserved and divergent lipocalin motifs (roman numbers) are given in black and red, respectively. The size of the lipocalin motif was determined by sequence alignment of VDE sequences and a representative group of lipocalin proteins. The C-terminal glutamic acid-rich domain indicates the percentage of Glu residues in this domain. B) Alignment of the N-terminal cysteine-rich domains of several plant and diatom VDEs. Also included is a sequence derived from the amoeba Acanthamoeba castellanii. C) Alignment of the lipocalin motifs I, II and III of several different lipocalin VDE, VDL and VDR proteins. The distance (in amino acids) between the three lipocalin motifs is also indicated. The lipocalin motif consensus sequences, as derived from kernel lipocalins (Flower, 1996), are indicated above the alignment and conserved motifs within the alignment are indicated in red. The abbreviations used are: Lip, lipocalin; TIL, temperature induced lipocalin; CHL, chloroplastic lipocalin; PRBR, plasma retinol-binding protein precursor; CC, crustacyanin; At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Dd, Dictyostelium discoideum; Gv, Gloeobacter violaceus; Hg, Homarus gammarus; Hs, Homo sapiens; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Pt, Phaeodactylum tricornutum; Py, Porphyra yezoensis; Ta, Triticum aestivum; Tp, Thalassiosira pseudonana; Vc, Vibrio cholerae.