Figure 1.
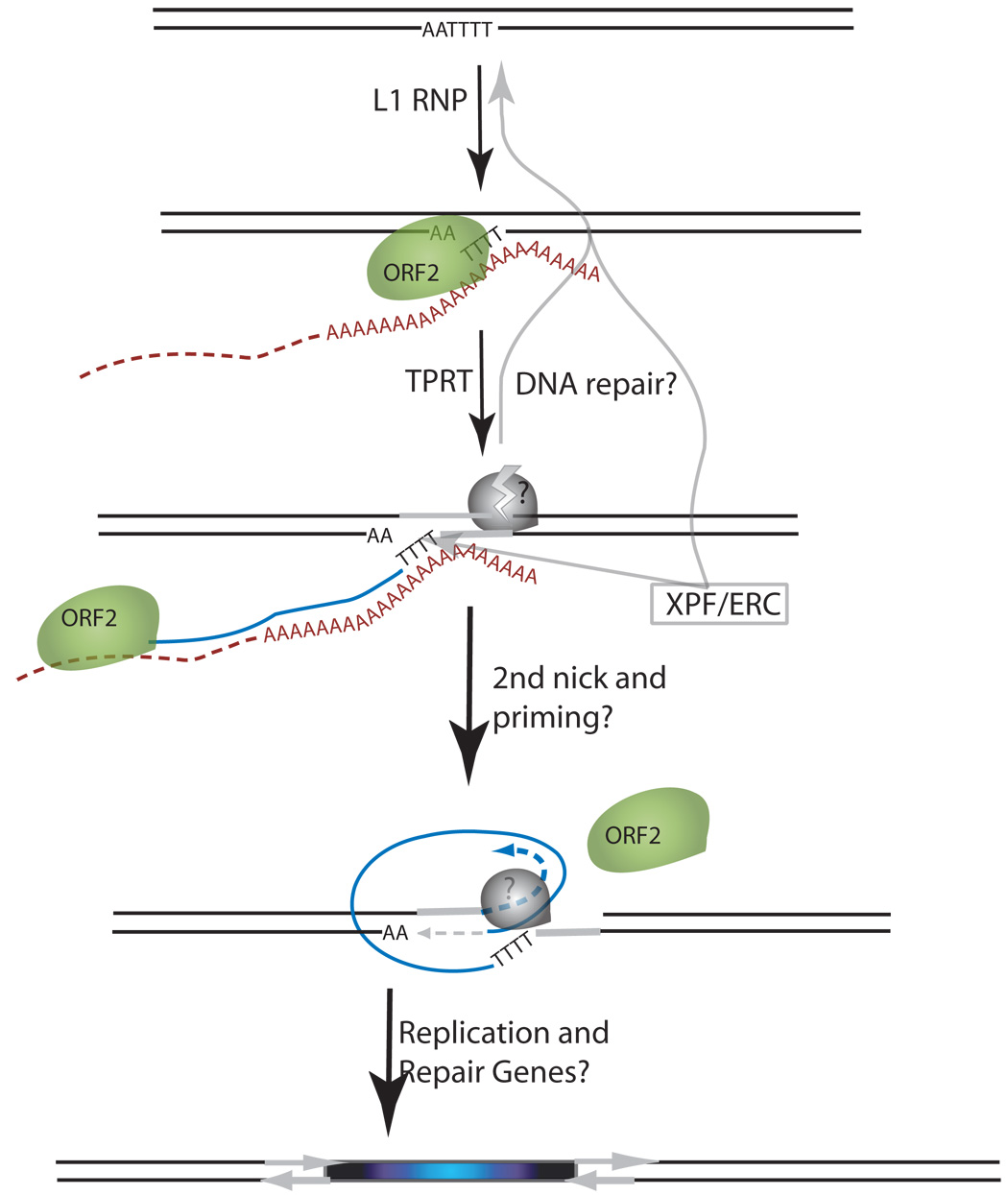
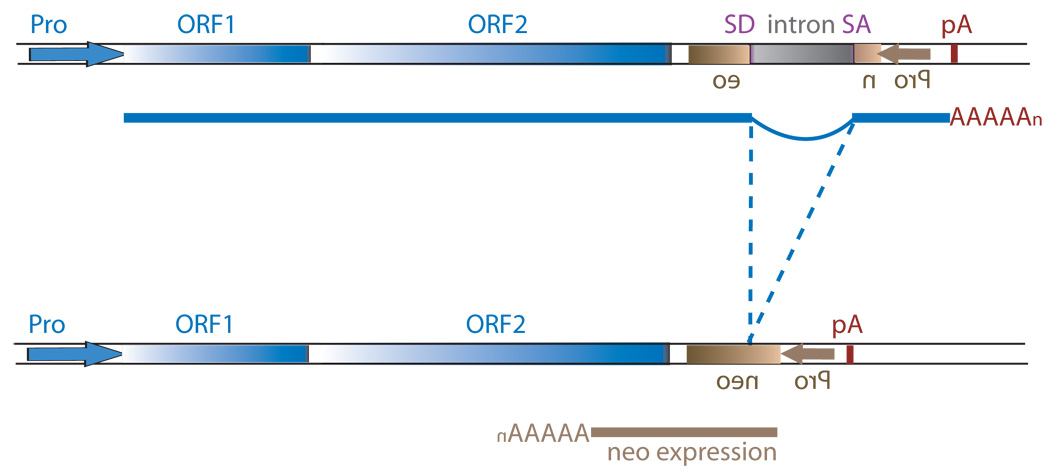
Schematic of the L1 TPRT reaction and retrotransposition assay. (A) A genomic site with a typical L1 endonuclease cleavage site (5’TTTTAA) (step 1) is exposed in the nucleus to the L1 RNP after the ORF2 cleaves the consensus site (step 2). The exposed T-rich region primes reverse transcription on the L1 mRNA polyA tail (step 3), and L1’s reverse transcriptase activity of ORF2 synthesizes cDNA (blue) forming a “flap intermediate” with a 3’ end. This intermediate is a known substrate for the ERCC1/XPF heterodimer. Processing by ERCC1/XPF is predicted to result in restoration of the original target DNA sequence. A nick occurs on the second strand via an unknown mechanism. The segment between the two nicks is highlighted in gray to illustrate the eventual formation of flanking direct repeats by the duplication of these segments. Second-strand synthesis is primed by microhomology-mediated priming which results in synthesis of a second copy of the gray segment (dotted arrow, step 4). Replication from the gray arrow completes synthesis across the 2nd strand of the cDNA creating a new L1 insert and completing synthesis across the other side of the direct repeat (step 5). (B) Schematic of the L1 retrotransposition assay. The L1.3 ORF1 and ORF2 are expressed by the CMV promoter without the L1 5’ UTR. The neomycin resistance gene is under the control of the SV40 promoter in the reverse orientation. The neoR coding sequence is interrupted by the human gamma-globin or the TNF intron which is spliced in the forward orientation. Subsequent integration via retrotransposition creates a neoR transgene that allows for colony growth under selection.
