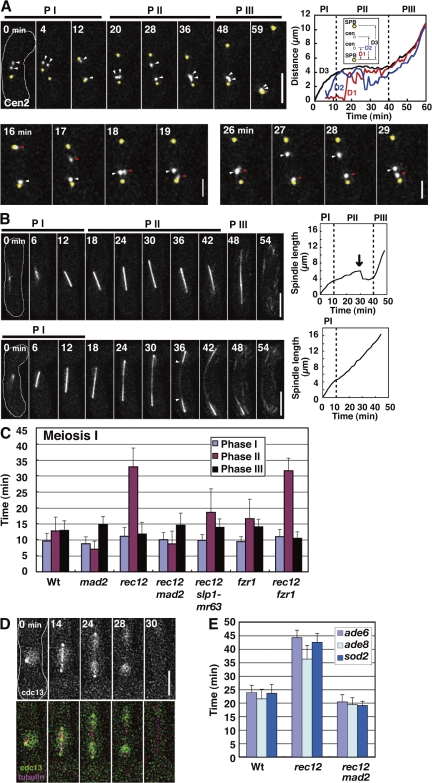
Figure 2.
Dynamics of the chromosome, the spindle, and Cdc13 at MI in rec12 mutant. (A) Behavior of homologous centromeres and the SPB at MI. Arrowheads indicate the homologous centromeres (cen2). Bottom panels highlight independent oscillations of the homologous centromeres (white and red arrowheads) at phase II. The graph shows changes in the SPB-cen (D1 and D2) and SPB-SPB (D3) distances. (B) Behavior of the MI spindle in rec12. The arrow in the top graph shows the sudden regression of the spindle. Arrowheads indicate the thin spindle midzone. Dotted lines in graphs show boundaries of the spindle phases. PI, phase I; PII, phase II; PIII, phase III. (C) Duration of spindle phases. (D) Dynamics of Cdc13-GFP at MI. (E) Timing of arm locus separation at MI. Bars show time of separation of the ade6, ade8, and sod2 loci after spindle formation. At least seven cells were examined for each locus. Wt, wild type. Error bars indicate standard deviation. Bars: (A, top, B, and D) 5 μm; (A, bottom) 2 μm.