Figure 2.
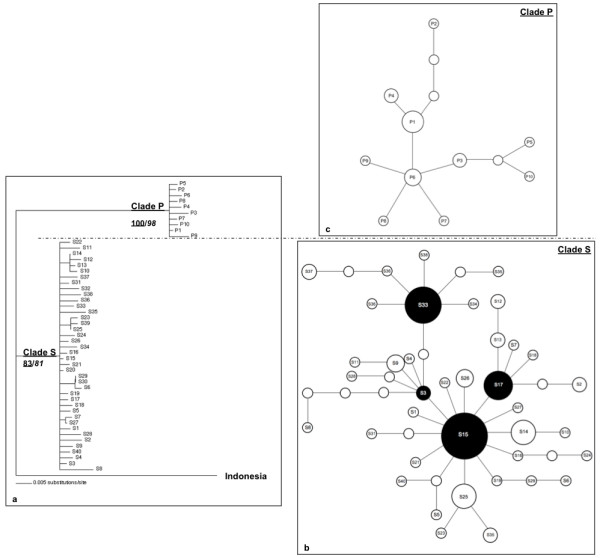
a) Neighbor joining phylogenetic tree of mtDNA sequences for M. schreibersii. Bootstrap support values for clades S and P are underlined for the neighbor joining phylogram and in italics for the maximum parsimony tree. A sample of M. schreibersii from Indonesia was used as outgroup. b) Statistical parsimony network of haplotypes (S1–S40) for clade S. The unlabeled circles indicate hypothetical haplotypes. The size of each circle is proportional to the frequency of the particular haplotype in the sample. c) The statistical parsimony network of haplotypes (P1–P10) for clade P. The unlabeled circles indicate hypothetical haplotypes. The size of each circle is proportional to the frequency of the particular haplotype in the sample.
