TABLE 2.
Comparison of models from TASSER_2.0, TASSER and PROSPECTOR_3.5
| 〈RMSD to the native〉, Å[SD*]
|
|||||||
|---|---|---|---|---|---|---|---|
| Num† | ID (%)‡ |
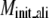 § §
|
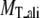 ¶ ¶
|
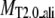 ‖ ‖
|
MT_all** | 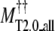 |
|
| Easy | 1802 | 19 | 5.60 [4.33] | 3.42 [2.55] | 3.27 [2.35] | 4.02 [2.80] | 3.86 [2.61] |
| Medium | 167 | 16 | 8.69 [5.16] | 5.39 [3.54] | 4.71 [2.77] | 5.82 [3.61] | 5.09 [2.87] |
| Hard | 622 | 13 | 11.87 [5.66] | 8.37 [4.57] | 7.69 [4.17] | 8.92 [4.48] | 8.24 [4.12] |
| All | 2591 | 17 | 7.30 [5.44] | 4.73 [3.84] | 4.42 [3.46] | 5.31 [3.92] | 4.99 [3.57] |
Standard deviation.
Number of target proteins in each category.
Average sequence identity of target-template sequences.
Average RMSD to the native structure of the initial PROSPECTOR_3.5 templates§, TASSER¶, and TASSER_2.0‖ models over the same aligned regions provided from PROSPECTOR_3.5.
Average RMSD to the native structure of TASSER** and TASSER_2.0†† models over the entire molecule.
