Figure 3.
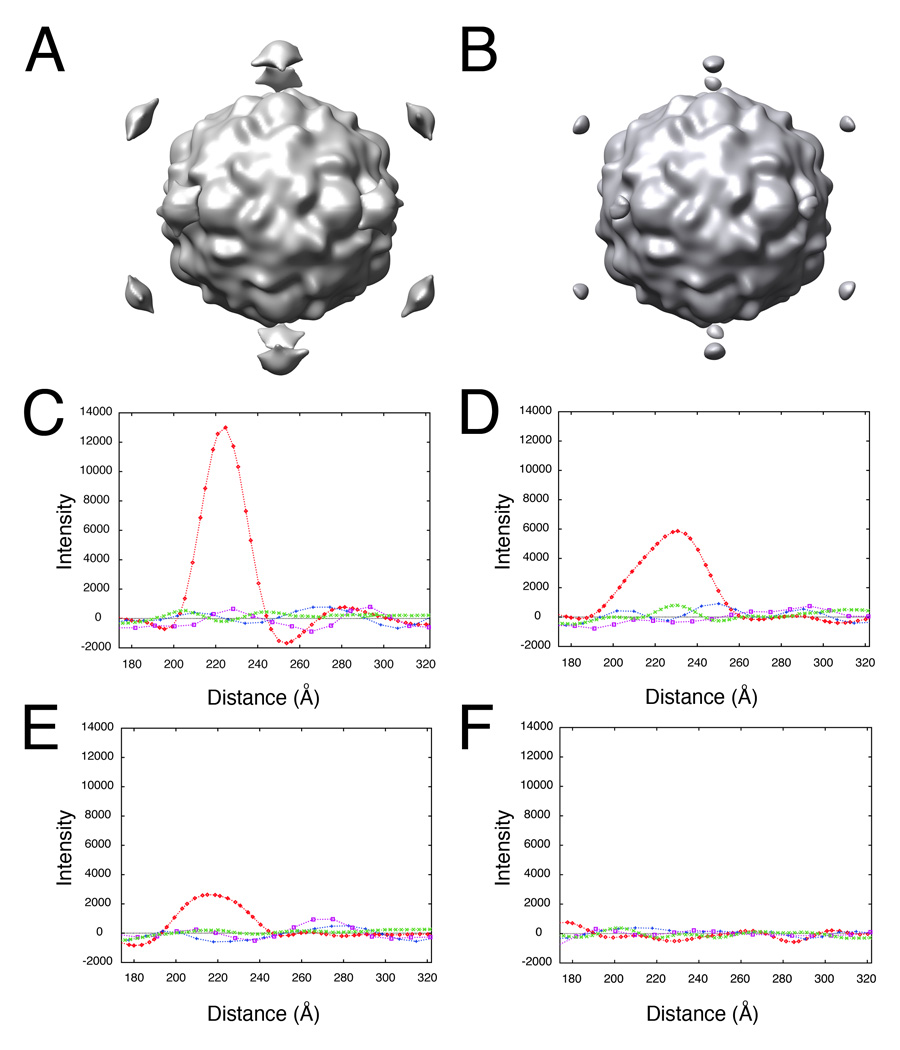
Determining the lower-order symmetry of the virus-receptor-liposome complex is a robust procedure that tolerates high levels of error in 2D spot placement. Random Gaussian error of 0 (A, C), 5 (D), 10 (B, E), and 45 (F) pixels deviation from the true membrane contact point were applied to the 2D spot coordinates of raw images. Icosahedral orientations for unspotted images, obtained using native virus as a reference, were used to compute 3D icosahedral reconstructions from these dotted 2D images (A, B). (C–F) Intensities in each reconstruction were sampled along the 5fold (red diamonds), 2fold (blue plusses), 3fold (purple squares), and quasi-3fold (green crosses) symmetry axes. Elevated density appearing approximately 220 Å from the center of the virus along a 5fold axis is evident, even with a 10 pixel (20.7 Å) RMS error in dot placement.
