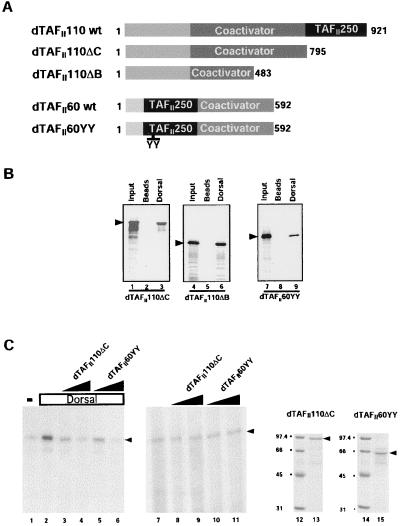
Figure 3.
(A) Schematic diagram of the domain structure of wild-type and mutant dTAFII110 and dTAFII60. The black boxed area represents the dTAFII250 interaction domain of each TAF. The dark gray area (marked coactivator) delineates the domains that are thought to interact with activator proteins. (B) TAFII110ΔC, TAFII110ΔB, and TAFII60YY retain the ability to bind selectively to Dorsal. Anti-FLAG antibody resin (lanes 2, 5, and 8) or beads loaded with Dorsal (lanes 3, 6, and 9) were incubated with [35S]methionine-labeled, in vitro-translated mutant TAFs indicated at the bottom of each panel. Protein complexes were separated by SDS/PAGE. Bound mutant TAFs were identified by autoradiography. Lanes 1, 4, and 7 represent 10% of the input material for each binding reaction. (C) Mutant dTAFII110 and dTAFII60 squelch activation by Dorsal but not basal transcription. Reactions containing the basal factors, RNA polII, 3 × DlTwiE1BCAT DNA template, and 0 ng of Dorsal (lane 1 and lanes 7–11) or 40 ng of Dorsal (lanes 2–6) were assayed in the presence of increasing amounts (30 ng or 100 ng) of either TAFII110ΔC (lanes 3 and 8 and lanes 4 and 9, respectively) or TAFII60YY (lanes 5 and 10 and lanes 6 and 11, respectively). Coomassie blue-stained SDS/PAGE gels of mutant TAFII110ΔC (lane 13) and TAFII60YY (lane 15) as well as molecular weight markers (lanes 12 and 14) are shown.