Fig. 6. Effect of μ2 amino acid Pro/Ser208 on complementation of M1-si-based reduction of infectious viral yields.
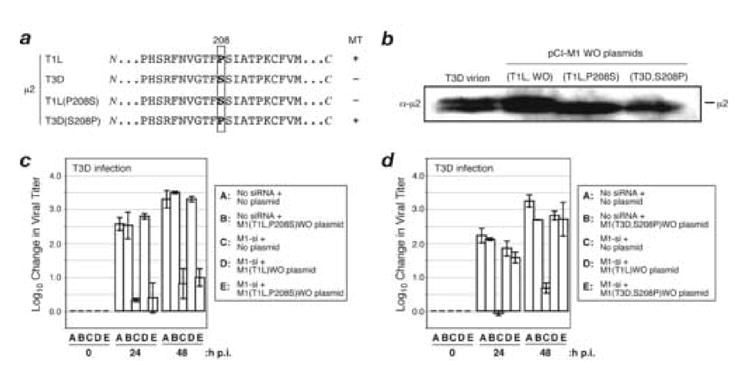
(A) Protein sequences for μ2 from reovirus strains T1L or T3D, or plasmids encoding T1L(P208S) or T3D(S208P) μ2. Box indicates the position of amino acid 208. T1L reovirus and plasmid encoding T3D(S208P) μ2 have a Pro at this position and encode a μ2 protein capable of strongly associating with microtubules (+); T3D reovirus and plasmid encoding T1L(P208S) μ2 have a Ser substituted at this position and weakly associate with microtubules (−). (B) CV-1 cells were transfected with 5 μg of either M1(T1L,P208S)WO or M1(T3D,S208P)WO rescue plasmids. The cells were incubated for an additional 24 h p.i., and cell-associated proteins were prepared for SDS-PAGE and immunoblotting. Protein was detected using specific antibodies against protein μ2. (C, D) CV-1 cells were transfected with no siRNA, rescue plasmid alone, M1-si alone, or M1-si in combination with a particular rescue plasmid, and at 24 h p.t. were infected with T3D reovirus at 10 PFU/cell. Cells were harvested at 0, 24, or 48 h p.i., and viral titers were determined by plaque assays. Log10 changes in viral titer relative to time 0 are indicated. Each bar represents the average obtained from three independent experiments, and error bars indicate the standard deviations.
