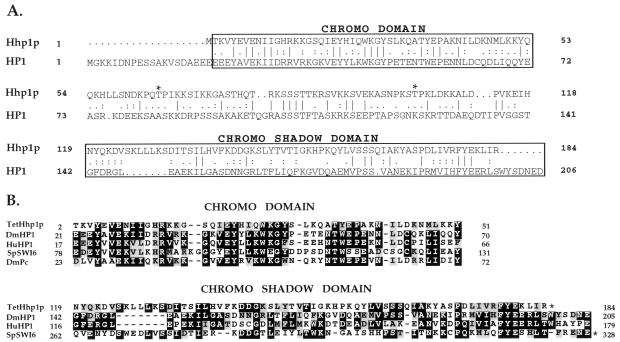
Figure 3.
Sequence alignment among Tetrahymena HHP1p, Drosophila HP1, and selected chromodomain family members (A and B). DmHP1, D. melanogaster (28); HuHP1, human (40); SpSWI6, Schizosaccharomyces pombe (35); DmPc, D. melanogaster (11). The alignment was made by the program clustal w (http://alfredo.wustl.edu/msa/clustal.cgi) and fine adjusted manually. The shading was then made by the program boxshade (http://ulrec3.unil.ch:80/software/BOX_form.html). Gaps in sequence alignments are indicated by dashes. Domain positions in the protein sequences are indicated by numbers; domains that end at the C terminus of the protein are marked with asterisks.