Figure 5.
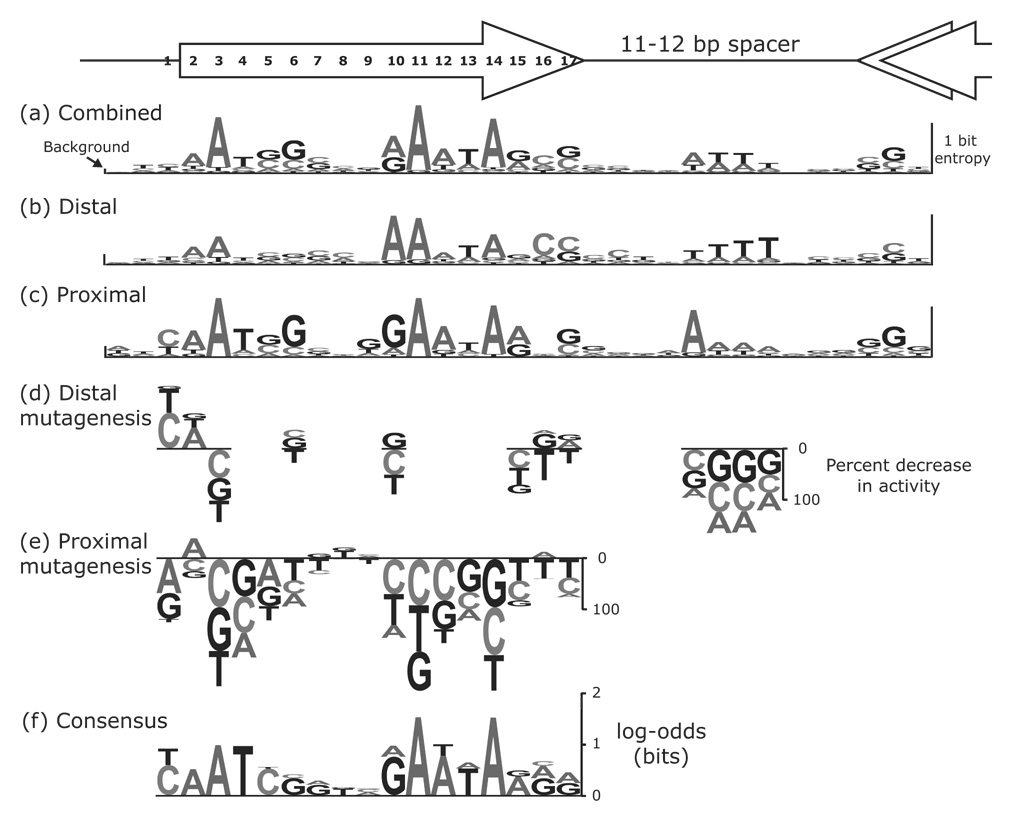
Comparison of the class 2 consensus sequence to the activities of fliAZY promoter mutants.Eighty-six proximal and distal FlhD4C2 sites were aligned to give the entropies in (a). The entropies for distal sites (b) or proximal sites (c) from an alignment of 30 “unidirectional” promoters are also displayed. The height of each WebLogo-like nucleotide stack49 corresponds to that position’s entropy,16 which is a measure of the conservation of nucleotides at that position. The frequency of each nucleotide in the alignment is reflected by the height of that nucleotide within the stack. On the left side of the figure, the height of the bar labeled background is twice the small sample correction as calculated by WebLogo.49 The small sample correction gives the average entropy value for a random alignment of sequences.50 All single base pair mutations at eight positions in the distal site (d), at four positions in the spacer (d), and at 17 positions in the proximal site (e) of the fliAZY promoter were constructed. The height of each letter corresponds to the percent change in promoter activity as quantified by β-galactosidase assays. Letters above or below the line indicate an increase or decrease, respectively, in promoter activity. (f) The measured activities for 17 positions in the distal and proximal sites were used as counts in order to generate a new consensus sequence. The activities for equivalent positions in the distal and proximal sites were combined, and the log-odds scores that are displayed were calculated using these counts. The log-odds scores are a measure of the conservation of nucleotides in the alignment.
