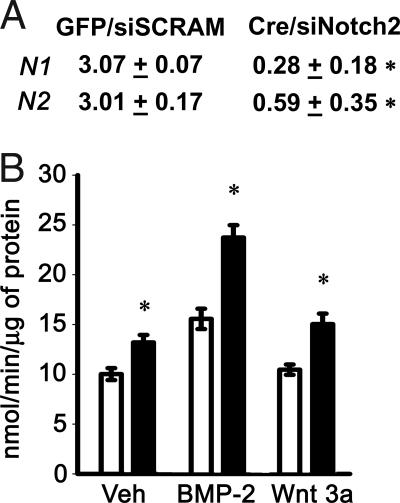
Figure 5.
Effect of Notch signaling inhibition on osteoblastic differentiation in calvarial osteoblasts harvested from notch1loxP/loxP mice transduced with Ad-CMV-GFP or Ad-CMV-Cre and transfected with Notch2 siRNA. Transduced osteoblasts were cultured to 70% confluence, and transfected with Notch2 (siNotch2) or control scrambled siRNA (siSCRAM). A, Deletion of Notch1 and down-regulation of Notch2 mRNA were documented in parallel cultures by real-time RT-PCR at the completion of the experiment. Data are expressed as notch1 (N1) and notch2 (N2) copy number, determined by real-time RT-PCR, corrected for gapdh expression. Values represent means ± sem for three observations. *, Significantly different from control cells, P < 0.05. B, Ad-CMV-GFP-transduced cells were transiently transfected with scrambled siRNA (white bars) and used as controls, whereas Ad-CMV-Cre-transduced cells were transiently transfected with notch2 siRNA (black bars). Twenty-four hours after transfection, cells were exposed to control medium (Veh), BMP-2 at 3 nm, or Wnt 3a at 2.7 nm and APA quantified in extracts from cells cultured for 72 h. APA is expressed as nanomoles of p-nitrophenol per minute per microgram of total protein. Bars, means ± sem for six observations. *, Significantly different from control cells, P < 0.05.