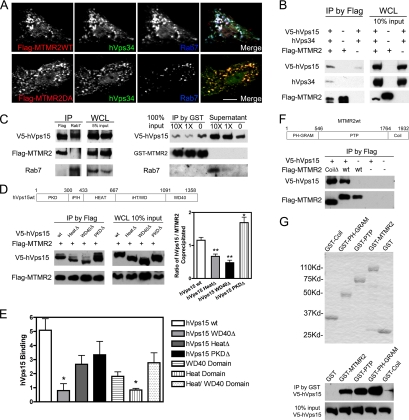
Figure 6.
The WD40 domain of hVps15 mediates exclusive binding of MTMR2 or Rab7 to the hVps34/hVps15 PI 3-kinase complex. (A) BHK cells overexpressing Flag-MTMR2wt or Flag-MTMR2D320A (catalytically inactive mutant), hVps34 and Rab7 were immunostained. Images of Flag-MTMR2 (red), hVps34 (green), and Rab7 (blue) are shown in gray-scale and color merge. Color merge shows colocalization (white) of MTMR2 with hVps34 on Rab7-positive late endosomes. Images were collected using 40× objective and scan zoom 1.0. Bar, 10 μm. (B) BHK cells overexpressing Flag-MTMR2, hVps34 and V5-hVps15 were processed for immunoprecipitation (IP) with an anti-Flag mAb. Western blot shows coprecipitated hVps34 or hVps15 (left panel) and expression levels of individual protein in whole cell lysates (WCL) for each sample with indicated input amounts (right panel). MTMR2 associates with the hVps34/hVps15 complex. (C) BHK cells overexpressing Rab7, Flag-MTMR2 and V5-hVps15 were processed for IP with anti-Rab7 or anti-Flag antibodies, respectively. Western blot shows coprecipitated hVps15 (left panel) and WCL (middle panel). MTMR2 and Rab7 form a mutually exclusive complex with hVps15. GST-MTMR2, 60 ng, immobilized on glutathione-Sepharose beads was incubated with identical aliquots of V5-hVps15–containing cell lysates. After washing, increasing concentrations of Rab7-containing cell lysates (0, 1×, 10×) were added for incubation. Western blot shows coprecipitated proteins on beads and proteins in the supernatant (right panel). Binding of MTMR2 or Rab7 to hVps15 is competitive. (D) BHK cells overexpressing full-length V5-hVps15 (wt) or domain deletion mutants (Δ) (indicated in top panel) and Flag-MTMR2 were processed for IP with an anti-Flag mAb. Western blot shows coprecipitated hVps15 (left panel) and WCL (middle panel). Right graph shows the ratio of coprecipitated hVps15 to MTMR2 in each sample quantified with chemiluminescent signal on immunoblots by densitometry (mean ± SEM, n = 4; unpaired t test, two-tailed *p < 0.05, **p < 0.01). hVps15 WD40Δ exhibits diminished complex formation with MTMR2. (E) Equimolar amounts of GST-MTMR2 immobilized on glutathione-Sepharose beads were incubated with in vitro–synthesized, 35S-l-methionine–labeled, full-length, hVps15 domain deletion mutants or individual hVps15 domains. Binding of the hVps15 proteins to GST-MTMR2 was quantified by phosphoimage analysis and binding was normalized relative to binding to GST (nonspecific binding equal to 1), which served as the negative control (mean ± SEM, n = 2, unpaired t test, two-tailed *p < 0.05). hVps15 WD40Δ exhibits diminished complex formation with MTMR2, but binding is reconstituted with a HEAT/WD40 domain. Representative binding data are shown in Supplementary Figure S6 online. (F) BHK cells overexpressing Flag-MTMR2wt or Flag-MTMR2 coli domain deletion mutant (CoilΔ) (indicated in top panel) with V5-hVps15 were processed for IP with an anti-Flag mAb. Western blot shows coprecipitated hVps15 (bottom panel). The MTMR2 coil domain is dispensable for binding to hVps15. (G) Equimolar amounts of full-length GST-MTMR2, GST-PH, GST-PTP, GST-Coil, or GST proteins were purified and confirmed with Coomassie staining (top panel). The GST fusion proteins immobilized on glutathione-Sepharose beads were incubated with equal amount V5-hVps15–containing cell lysates. Western blot shows coprecipitated hVps15 (bottom panel). Both the PH and PTP domains of MTMR2 bind to hVps15 independently.