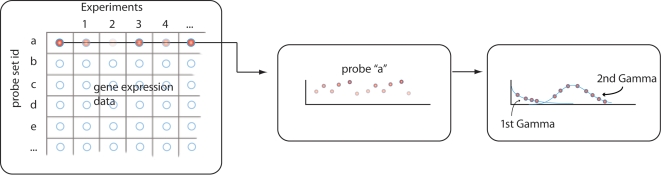
Figure 1. Data handling in the mixture algorithm.
First, gene expression data from a set of gene expression experiments is collected. The matrix in the figure shows rows probe set, where every row is a single probe set, and every column is a different hybridization experiment. This could be, for example, Affymetrix microarray experiments, where each column is a different patient. We then look at the data probe-by-probe. For example, we follow probe “a” in the figure and look at the expression levels for this probe, across all samples in the set of gene expression experiments. Each probe will have data from the entire collection of experiments. For the specific probe “a”, we fit the set of expression measurements into two gamma distributions, one representing the “down” state and one representing the “up” state. Each data point is then computationally associated with a probability of being either under the first Gamma distribution (which would mean the gene associated with the probe, for the specific sample, is at a “down” state) or with the second Gamma distribution (which would mean the gene associated with the probe, for the specific sample, is at an “up” state). We iterate the procedure across the entire probe-set, to tag every gene across the microarray with its probability of being “up” or “down”.