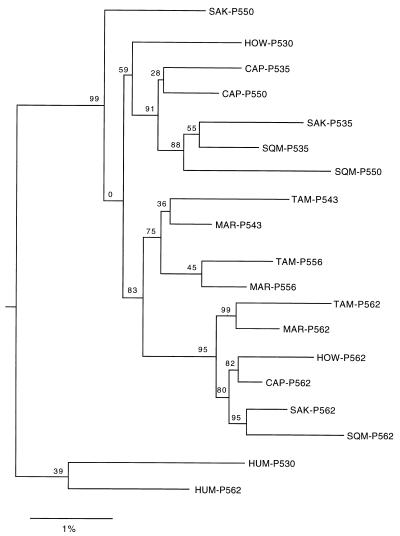
Figure 4.
Neighbor-joining tree derived from an analysis of exon 3, 4, and 5 sequences. The distances were computed by using the method of W.-H.L. (27), with weights of 80% and 20% for nonsynonymous and synonymous substitutions, respectively. The number at each node denotes the percent of 500 bootstrap replicates that supported the subset of sequences. The galago sequences of Zhou et al. (30) were used to root the tree (CAP, capuchin; HUM, human; HOW, howler monkey; MAR, marmoset; SAK, saki monkey; SQM, squirrel monkey; TAM, tamarin).