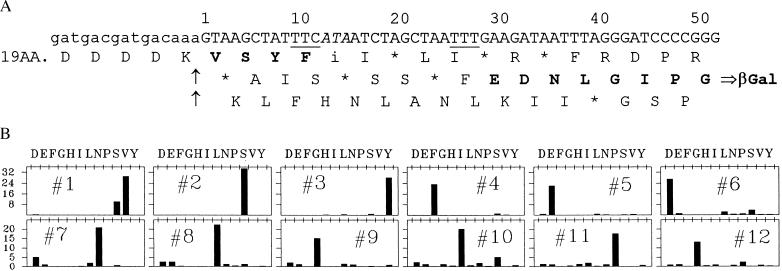
Figure 2.
Nucleotide sequence of the coding strand and amino acid sequence of the protein encoded by construct L-16 (TTT) in the relevant region. (A) The coding sequence is shown numbered from the G in the first position of the codon for the valine on the C-terminal side of the enterokinase cut site, shown by arrows. The 0-frame sequence on the N-terminal side of the cut site is indicated schematically. Predicted translation products in all three reading frames are shown, aligned under their corresponding triplets, with termination at nonsense triplets indicated by an ∗. The shifty sequence T TTC ATA is at positions 9–15. Amino acids in boldface are those found in the protein made in isoleucine-tRNA limited cells. The phenylalanine codons at the takeoff and landing sites are underlined. The hungry isoleucine codon is italicized. (B) Cells carrying L-16 (TTT) (see Table 1) were grown into logarithmic phase, the lac promoter was induced, and isoleucyl-tRNA synthetase was partly inhibited by addition of 72 μg/ml ILHX. After two to three doublings of OD, the cells were harvested and β-glactosidase was immunopurified for amino acid sequence analysis as described (9). The β-galactosidase immunoprecipitate was dispersed in 1× EKMax buffer (50 mM Tris⋅HCl, pH 8.0, 1 mM CaCl2, 0.1% Tween 20) and treated with enterokinase for 2 hr at 37°C (9). The cleavage mixture was run on 6% PAGE gels and electroblotted to poly(vinylidene difluoride) membrane. The results of Edman sequence analysis (performed at Southwest Scientific Resources) are shown for each successive cycle, with lag-corrected pmol on the y axis, and the different amino acids on the x axis. Similar results were obtained with another sample sequenced at the Stanford Protein and Nucleic Acid Facility.