Figure 2.
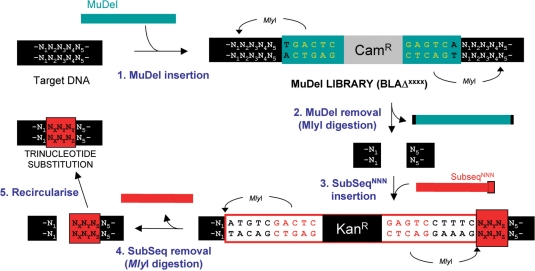
Mechanism of TriNEx process. Step 1. Two MlyI recognition sites (5′GAGTC(N)5↓) were placed 1 bp from each end of MuDel, 1 bp away from the site of transposon insertion. MlyI cuts 5 bp outside its recognition sequence to generate a blunt end. Insertion of MuDel results in the duplication of 5 bp (N1N2N3N4N5) of the target gene at the insertion point (23,24). Step 2. Digestion with MlyI removes MuDel together with 8 bp of the target DNA (4 bp at each end). This equates to removal of a contiguous 3 bp sequence from the starting target gene (N2N3N4). Step 3. SubSeqNNN is then ligated into the gap vacated by MuDel. SubSeqNNN also contains two MlyI recognition sites strategically placed towards the ends of the cassette. One site is located so that MlyI will cut at the exact point where the target DNA joins SubSeqNNN. The second site will cut 3 bp into SubSeqNNN so donating 3 bp (NXNYNZ) to the target DNA. Step 4. Digestion with MlyI removes SubSeq but with 3 bp of its sequence now replacing the 3 bp deleted from the target gene. Step 5. Intramolecular ligation reforms the target gene but with one contiguous trinucleotide sequence replaced with another.