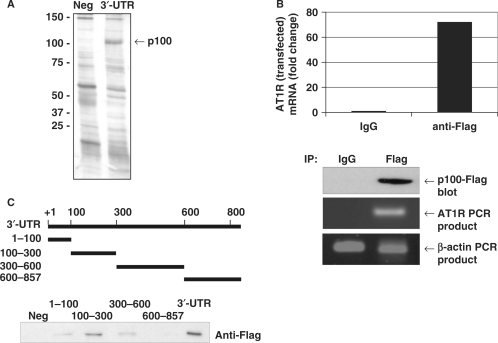
Figure 1.
p100 interacts with AT1R 3′-UTR. (A) Search for AT1R 3′-UTR-binding proteins using affinity purification. The details of the purification are given under ‘Experimental Procedures’. A negative control probe (Neg) consisting of the segment of luciferase coding region and a probe corresponding to nucleotides 1–857 of AT1R 3′-UTR (3′-UTR) were incubated with HEK293 cell lysates and run into SDS–PAGE, which was then silver-stained. The differentially expressed 100 kDa protein was analysed by mass spectrometry. (B) Amplification of AT1R mRNA from p100 IPs. p100-Flag was immunoprecipitated with anti-Flag antibody or control IgG. RNA was extracted from the IPs, subjected to RT–PCR, followed by quantitative PCR analysis of AT1R and β-actin (loading control) mRNAs. The AT1R mRNA levels were normalized to β-actin mRNA levels. Results are expressed as a fold change compared to AT1R mRNA precipitated by IgG. Lower panels show control anti-Flag western blot and agarose gels with AT1R and β-actin PCR products. (C) p100 interacts with 100–300 of AT1R 3′-UTR. A schematic representation of different 3′-UTR transcripts used as probes in affinity purification reactions is shown. Probes were incubated with p100-Flag protein containing cell lysates. The results are shown as western blot demonstrating bound Flag-p100.