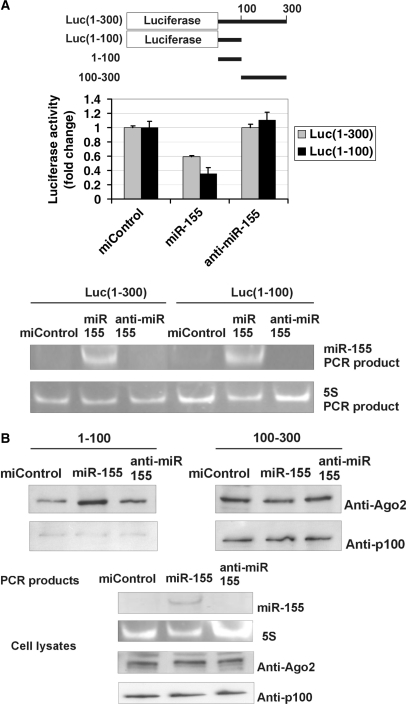
Figure 4.
MiR-155 does not require p100 to function. (A) MiR-155 inhibits the activity of Luc(1–300) and Luc(1–100) constructs. A schematic representation of the luciferase constructs used in Figure 4A and 3′-UTR probes used in Figure 4B is shown. COS-1 cells were transfected with a Luc(1–300) construct with 1–300 nucleotides of 3′-UTR, or Luc(1–100) construct with 1–100 nucleotides of AT1R 3′-UTR and co-transfected with 30 nM of negative control miRNA (miControl), miR-155 or anti-miR-155 together with plasmid expressing renilla luciferase. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The mean luciferase value from miControl-transfected cells was set to one. Below are the agarose gel figures showing the expression of miR-155 and 5S detected by PCR. (B) MiR-155 specifically increases the binding of Ago2 to 1–100 region of 3′-UTR, but not to 100–300 region. MiR-155 has no effect on p100 binding to either probe. COS-1 cells were transfected with miControl, miR-155 or anti-miR-155. Corresponding cell lysates were affinity purified by probes spanning 1–100 or 100–300 nucleotides of the 3′-UTR, as indicated. The binding results were analysed by western blot analysis with anti-Ago2 or anti-p100 antibodies (upper two panels). Below are the control gels illustrating the expression levels of miR-155 and 5S (PCR products in agarose gels), and Ago2 and p100 (western blots) in cell lysates.