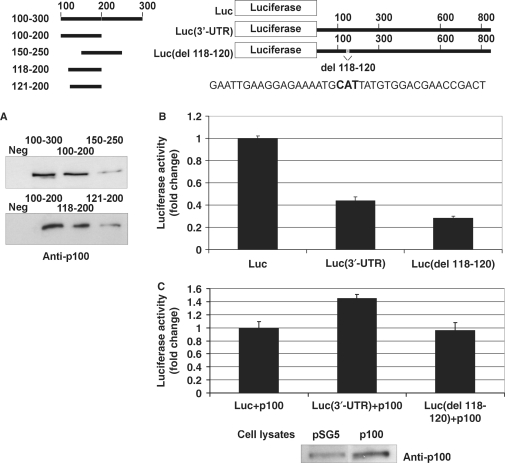
Figure 7.
Mapping and deletion of the p100-binding site. A schematic of the probes used in the affinity purification and luciferase constructs used in luciferase assays is shown. The schematic also shows the sequence around 118–120 (118–120 shown in bold). (A) Nucleotides 118–120 of AT1R 3′-UTR are critical for p100 interaction. A negative control probe (from luciferase coding region; Neg) or different 3′-UTR probes were incubated with cell lysates and the binding products were analysed by western blot with anti-p100 antibodies. (B) Deletions of p100-binding site lead to reduced luciferase activity. COS-1 cells were transfected either with a luciferase coding region only (Luc), with Luc(3′-UTR), or with Luc(del 118–120) constructs, as indicated, and co-transfected with renilla luciferase. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The luciferase activity corresponding to transfection with Luc construct was set to one. (C) Deletion of p100 binding site (del 118–120). COS-1 cells were transfected with the same set of luciferase constructs as in (B), and co-transfected with empty pSG5 vector, or pSG5 plasmid expressing p100-Flag, together with renilla luciferase, as indicated. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The value corresponding to transfection with Luc construct together with empty pSG5 vector was set to one. Below is the representative control western blot of p100 expression.