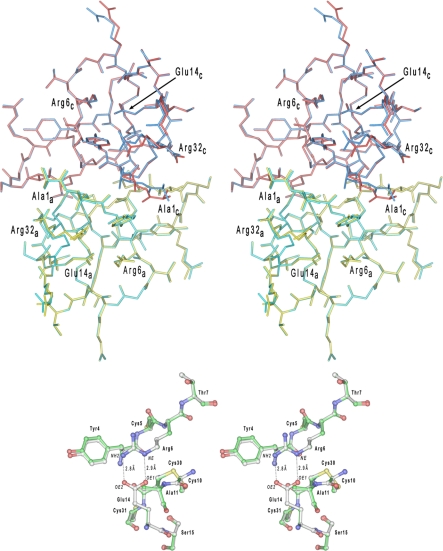
FIGURE 5.
Superposition of HD5 dimers in shown in a stereo representation. In the upper panel, a dimer of HD5 colored in yellow and red represents the monomers A and C in the previously determined crystal structure of this defensin and deposited in the Protein Data Bank under the code 1zmp. The second dimer (colored in cyan and blue) is composed of monomers A and C from the current structure. The N- and C-terminal residues, as well as Arg6 and Glu14 (forming the salt bridge), are labeled for both monomers. Both structures are virtually indistinguishable (root mean square deviation for the 64 equivalent Cα-atoms is 0.088 Å) with slight deviations found only for few highly flexible side chains. A region of the Arg6–Glu14 salt bridge is shown in the lower panel for both HD5 structures superimposed. The carbon atoms in the “1zmp” model are painted white, whereas in the current model, they are shown in green.