Abstract
Few genes in the divergent eukaryote Trichomonas vaginalis have introns, despite the unusually large gene repertoire of this human-infective parasite. These introns are characterized by extended conserved regulatory motifs at the 5′ and 3′ boundaries, a feature shared with another divergent eukaryote, Giardia lamblia, but not with metazoan introns. This unusual characteristic of T. vaginalis introns led us to examine spliceosomal small nuclear RNAs (snRNAs) predicted to mediate splicing reactions via interaction with intron motifs. Here we identify T. vaginalis U1, U2, U4, U5, and U6 snRNAs, present predictions of their secondary structures, and provide evidence for interaction between the U2/U6 snRNA complex and a T. vaginalis intron. Structural models predict that T. vaginalis snRNAs contain conserved sequences and motifs similar to those found in other examined eukaryotes. These data indicate that mechanisms of intron recognition as well as coordination of the two catalytic steps of splicing have been conserved throughout eukaryotic evolution. Unexpectedly, we found that T. vaginalis spliceosomal snRNAs lack the 5′ trimethylguanosine cap typical of snRNAs and appear to possess unmodified 5′ ends. Despite the lack of a cap structure, U1, U2, U4, and U5 genes are transcribed by RNA polymerase II, whereas the U6 gene is transcribed by RNA polymerase III.
Keywords: Trichomonas vaginalis, splicing, snRNA, capping
INTRODUCTION
RNA splicing, an essential step of mRNA maturation in eukaryotic genes containing introns, is catalyzed by a large ribonucleoprotein particle, the spliceosome (Burge et al. 1999). This dynamic particle is composed of U1, U2, U4, U5, and U6 small nuclear RNAs (snRNAs) and hundreds of proteins (Nilsen 2003). Spliceosomes have been best characterized in yeast and mammals, where well-established in vitro splicing systems are available (Nilsen 1998; Burge et al. 1999). Although spliceosomal protein components may vary between organisms and even within different tissues of a single organism, all spliceosomes share a conserved core of proteins and the five snRNAs (Burge et al. 1999).
Spliceosomal introns have been found in all examined eukaryotes (Vanacova et al. 2005), but are absent in eubacterial and archeal genomes. The prevalence and number of introns vary greatly in unicellular eukaryotes (Gardner et al. 2002; Abrahamsen et al. 2004; El-Sayed et al. 2005; Loftus et al. 2005; Carlton et al. 2007). For example, conventional cis introns are widespread in apicomplexan parasites (Gardner et al. 2002) and Entamoeba (Loftus et al. 2005; Davis et al. 2007), but are found only infrequently in trypanosomes (El-Sayed et al. 2005) and in the diplomonad Giardia lamblia (Nixon et al. 2002; Morrison et al. 2007). Euglenoids, relatives of trypanosomes, contain many cis introns, but also have a subset of unconventional introns that are excised by a spliceosome-independent mechanism (Ebel et al. 1999). Although the unicellular eukaryote Trichomonas vaginalis has an unusually large repertoire of genes, only ∼65 of these genes appear to have an intron (Vanacova et al. 2005; Carlton et al. 2007).
Conserved canonical 5′ splice site (SS), 3′SS, and branch-site signals are present in T. vaginalis introns; however, the unusually short branch site–3′SS distance (7 nt) creates a highly conserved 12-nt motif (5′-ACTAACACACAG/-3′) encompassing the branch-site adenosine (underlined) and the 3′SS (/), which is required for splicing (Vanacova et al. 2005). To further investigate RNA structural requirements for splicing in this divergent eukaryote, here, we have identified and characterized T. vaginalis snRNAs.
Spliceosomal snRNAs interact with conserved intron motifs, positioning the pre-mRNA in the complex and directing splicing specificity (Ares and Weiser 1995). Studies in yeast and metazoa have demonstrated that base pairing between snRNAs and intronic 5′SS, 3′SS, and branch site also provide a scaffold for spliceosomal assembly and catalysis (Nilsen 1998). First, the 5′SS and branch-site base pair with U1 and U2 snRNAs, respectively, followed by the entry of a U4/U6.U5 tri-snRNP complex (Burge et al. 1999). Subsequent conformational changes result in the replacement of U1 snRNA by U6 snRNA at the 5′SS, and unwinding of the U4–U6 snRNA complex. Base pairing between U6 and U2 snRNAs facilitates juxtaposition of the 5′SS and branch site at the catalytic center, allowing for the first transesterification step. In addition to positioning critical splicing motifs, U6 snRNA is thought to coordinate divalent cations needed for catalysis (Yean et al. 2000).
Spliceosomal snRNAs have been proposed to be the evolutionary descendents of self-splicing introns (Sharp 1985). The U2–U6 complex reveals structural similarity to a self-splicing group II intron-like domain, consistent with derivation of these snRNAs from this ancestral ribozyme. Furthermore, the human U2–U6 minimal catalytic domain has been shown to form a covalent product with a branch-site RNA in a reaction that resembles the first step of splicing catalysis (Valadkhan and Manley 2001). The U2–U6 core domain is structurally conserved and allows for the autonomous formation of a four-way helical junction (Valadkhan and Manley 2000; Sashital et al. 2004; Valadkhan 2005) that appears to be required for splicing in both yeast and mammals.
snRNAs are transcribed by RNA polymerase (RNAP) II, with the exception of U6 snRNA, which is a RNAP III transcript (Hernandez 2001). Trypanosomes break this rule by transcribing all of their snRNAs using RNAP III and an upstream inverted tRNA gene as promoter (Fantoni et al. 1994). As is typical of RNAP II transcripts, U1, U2, U4, and U5 snRNAs in yeast and mammals acquire a m7G cap cotranscriptionally. This cap is subsequently hypermethylated, creating a m2,2,7G (trimethylguanosine; TMG) cap structure (Busch et al. 1982). Hypermethylation of snRNA cap structures occurs in the cytoplasm in humans and in the nucleolus in yeast. The biological significance of the distinctive 5′-cap structure found in snRNAs is unclear, but it has been suggested to play roles in snRNA compartmentalization and/or transport (Huber et al. 2002; Mouaikel et al. 2002; Narayanan et al. 2003).
Our previous work revealed that introns in T. vaginalis and G. lamblia are similar to one another yet divergent from most other eukaryotic introns, in that they contain a conserved 12-nt motif encompassing the branch site and 3′SS (Vanacova et al. 2005). This motif was shown to be critical for splicing; however, the snRNAs predicted to interact with the motif were not identified in either T. vaginalis or G. lamblia. To this end, we have now identified and characterized all five T. vaginalis spliceosomal snRNAs.
RESULTS
Identification of putative spliceosomal snRNAs in Trichomonas vaginalis
Introns were recently identified in ∼65 T. vaginalis genes (Vanacova et al. 2005; Carlton et al. 2007); however, little is known about the splicing machinery in this highly divergent eukaryote (Baldauf 2003). As a step toward characterizing splicing mechanisms in this unicellular parasite, we searched for the presence of evolutionarily conserved spliceosomal snRNAs. Short conserved regions in the spliceosomal snRNAs were used to screen the 7X coverage T. vaginalis genome database (Carlton et al. 2007). The sequence of U2 snRNA (CAAGTGTAGTATCTG; position nt 28 to nt 42 in human U2 snRNA) known to interact with the intron branch site, and a phylogenetically conserved sequence comprising the apical loop of U5 snRNA helix I (TTCGCCTTTTACTA, position nt 34 to nt 47 in human U5 snRNA) were utilized in Blastn searches. These revealed a number of candidate snRNA genes that were subsequently analyzed using structural prediction software RNAfold (Hofacker 2003). Sequences immediately upstream and downstream from the target sequence were tested for the presence of predicted secondary structures and conserved nucleotides in these regions. This approach resulted in the identification of a single gene encoding each of the putative U2 and U5 snRNAs in the T. vaginalis genome.
Using INFERNAL software and multiple alignments of U6 snRNA databases to screen the T. vaginalis genome, a putative U6 snRNA gene was also found. U6 snRNA forms intermolecular complexes with both U2 and U4 snRNAs, essential for spliceosome assembly and function (Brow and Guthrie 1988; Wu and Manley 1991; Madhani and Guthrie 1992; Sun and Manley 1995). Using a short region of the identified U6 gene (GTCCTTGGACAAAGA, nt 71 to nt 85) predicted to base pair with U4 snRNA, we then identified a candidate U4 snRNA gene utilizing Blastn and the RNA-folding approaches described above for identification of U2 and U5 snRNAs.
A Blastn search for U1 snRNA T. vaginalis gene using a conserved sequence comprising the U1A binding site (CCATTGCACTCCGG, position nt 63 to nt 76 in human U1 snRNA) was unsuccessful. Thus, we turned to a functional approach to identity this snRNA. We first identified a gene with similarity to human U1-70K (e-value of 2e−13 by Blastp) and a central RRM domain (Nelissen et al. 1994) in the T. vaginalis genome. This protein, tagged with streptavidin binding protein (SBP) to allow for affinity purification on streptavidin sepharose, was expressed in T. vaginalis transfectants and subsequently isolated from nuclear extracts. Bound RNA (100–200 nt) was extracted from this material, cloned, and sequenced, allowing for identification of U1 snRNA.
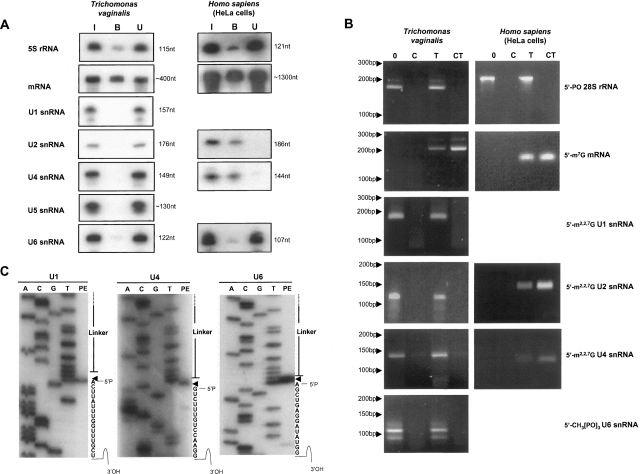
Subsequent to identifying the T. vaginalis snRNA, a BLASTn search of the genome database using each individual snRNA revealed only one gene copy for each of the five putative snRNA. However, given the possibility of sequence gaps, nucleotide divergence and loss of sequence data during the assembly of this large, repetitive genome (Carlton et al. 2007), it is possible that additional undetected genes are present. Northern analysis confirmed the expression of the five snRNAs identified (Fig. 4, below; data not shown) and their 5′ and 3′ ends were mapped by RNA ligase-mediated 5′ end rapid amplification of cDNA (RLM-5′ RACE), 3′ RACE, and DNA sequencing (Fig. 4, below; data not shown) except for the 5′ end of U5 snRNA, which was mapped by primer extension (data not shown).
FIGURE 4.
Comparative analysis of snRNA 5′-termini. (A) Anti-TMG immunoprecipitation of total RNA isolated from T. vaginalis (left) and human (right). Collected fractions: (I) input RNA, (B) bound RNA, and (U) unbound RNA were subjected to gel fractionation, blotting, and hybridization to specific probes. Probes (right) and RNA size (left) are indicated. Replica blots from the same immunoprecipitation assay were used. (B) Evaluation of snRNA 5′-ends by RNA Ligase-Mediated Rapid Amplification of cDNA 5′-Ends (RLM-5′RACE). T. vaginalis RNA and human RNA (control) analyses are shown at left and right, respectively. RNAs examined are listed (right) and size markers are indicated (left). Treatments are as follows: (0) untreated; (C) calf intestinal alkaline phosphatase (CIP) treatment only; (T) tobacco acidic pyrophosphatase (TAP) treatment only; (CT) CIP treatment followed by TAP treatment. Controls were a noncapped RNA species (28S rRNA) and a m7G capped mRNA (T. vaginalis ferredoxin and human GAPDH mRNAs). Targets (right) and DNA size (left) are indicated. (C) Primer extension analysis of T. vaginalis U4, U5, and U6 snRNAs subsequent to RLM-5′RACE analyses. Products of primer extension analyses (PE), flanked by sequence reactions (A, C, G, T) for identification of 5′ ends, are shown. Arrowheads mark the single 5′ end nucleotide of each snRNA and correspond to 5′ ends detected using untreated samples.
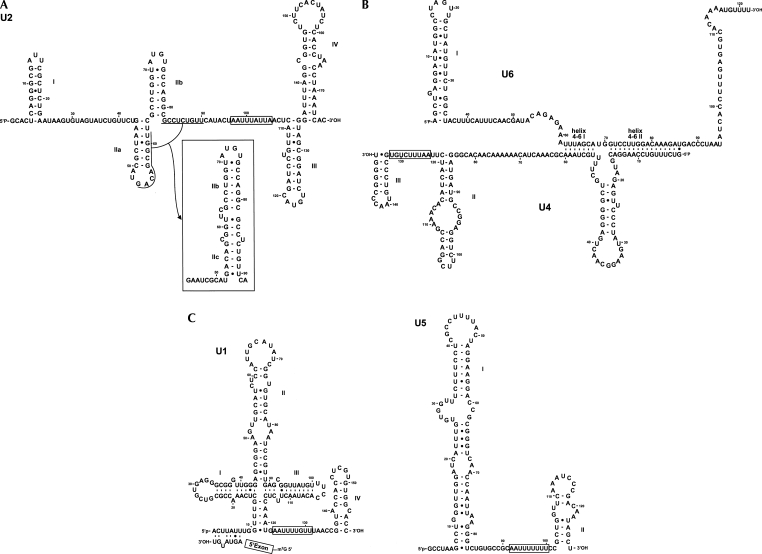
Sequence and secondary structure comparisons of the full-length snRNAs revealed each to be similar to other eukaryotic snRNA homologs (Figs. 1, 2) (Burge et al. 1999). The putative T. vaginalis U2 snRNA was found to possess all five conserved loops (I, IIa, IIb, III, and IV) with interspersion and length similar to those found in U2 of other organisms (Fig. 1A). T. vaginalis U2 snRNA has extensive primary sequence conservation at the 5′ end (Supplemental Fig. 1B), allowing formation of stem–loops I, IIa, and IIb (Fig. 1A). Interestingly, the loop and nucleotides at the 3′ side of helix IIa are complementary to a single-stranded region downstream from helix IIb (Fig. 1A,B), and thus can form stem IIc (Ares and Weiser 1995). Conformational changes between the phylogenetically conserved IIa and IIc forms are implicated in spliceosomal transitions (Hilliker et al. 2007; Perriman and Ares 2007). The putative U4 snRNA can be base paired with the putative U6 snRNA, forming two conserved helices separated by an intramolecular stem–loop. Although the Y-shaped U4–U6 interaction domain is conserved (Fig. 1B), the primary sequence of U4 snRNA is not (Supplemental Fig. 1C). Nevertheless, stem–loops II and III, with the uridine-rich Sm-binding site in between, can be formed in the U4 snRNA (Fig. 1B). The short U4–U6 stem I observed in T. vaginalis may be compensated by a longer stable U4–U6 stem II. The latter does not contain any disruption of Watson-Crick base pair thought to be important in balancing stability during spliceosomal structural transitions in vertebrates (Ares and Weiser 1995).
FIGURE 1.
Sequence and predicted secondary structures of T. vaginalis U1, U2, U4, U5, and U6 spliceosomal snRNAs. Nucleotides are numbered from 5′ to 3′ and putative Sm binding sites are boxed. Helices are indicated in roman numerals. (A) Predicted secondary structure of U2 snRNA; conserved loops are indicated in roman numerals. Sequence underlined shows potential base-pair interactions for formation of helix IIc as indicated in the inset. (B) Predicted secondary structures of U4 and U6 snRNAs, as indicated. U4/U6 intermolecular helices are marked as 4–6 I and 4–6 II. (C) Predicted secondary structure of U1 (left) and U5 (right) snRNAs. Interaction of 5′SS intron sequence from poly(A) polymerase pre-mRNA (Vanacova et al. 2005) with U1 snRNA is shown.
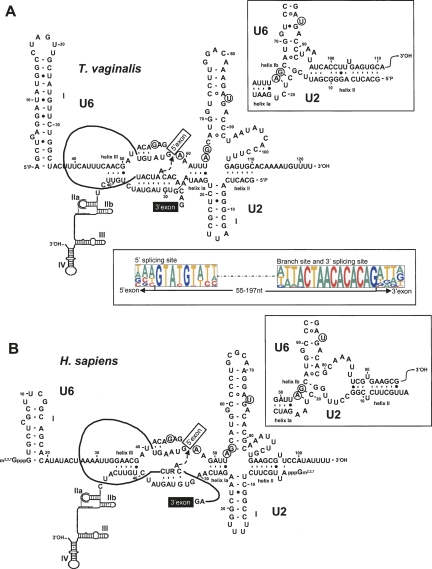
FIGURE 2.
Comparison of U2/U6 snRNA interaction models for T. vaginalis (A) and Homo sapiens (B). Spliceosomal T. vaginalis U2–U6 interactions are modeled after those proposed for yeast and mammalian homologs. Nucleotides proposed to be involved in coordination of metal ions required for catalysis (Fabrizio and Abelson 1992; Yu et al. 1995) are circled. The numbering of nucleotides and helices are indicated. The intron sequence from poly(A) polymerase pre-mRNA (Vanacova et al. 2005) and generic splice site and branch site sequences are incorporated in the T. vaginalis and the H. sapiens models, respectively. The U2/U6 snRNA conformation that allows for an extended U6 intramolecular stem–loop (ISL) proposed to form during the first step of catalysis is shown on the left; the inset on the right shows the proposed second step conformation that allows for formation of helix Ia and Ib. The nucleophilic attack by the branch-site adenosine during the first step of splicing is marked by an arrow. A consensus of the 5′SS, branch site and the 3′SS in identified T. vaginalis introns (Carlton et al. 2007) are shown below the T. vaginalis U2/U6 structure.
T. vaginalis U1 snRNA has a predicted secondary structure (Fig. 1C) more similar to human U1 snRNA than its counterpart in Saccharomyces cerevisiae U1 (Kretzner et al. 1987). The parasite U1 snRNA also exhibits a phylogenetically conserved region that could potentially interact with the 5′SS. This sequence ACUUAUU is almost identical to that found in metazoan U1 snRNAs (ACUUACC) (Supplemental Fig. 1A). Although the last two positions (underlined) differ between T. vaginalis and metazoan U1, these should still interact with 5′SS positions (Fig. 1C). This finding indicates that recognition of the 5′SS, an important step in intron recognition, is conserved throughout eukaryotic evolution. As we have previously reported (Vanacova et al. 2005), the 5′SS of T. vaginalis intron is more conserved than typically found in metazoan introns (Fig. 2A). The T. vaginalis U1 snRNA helices are also more highly conserved in comparison with those of metazoa U1 snRNAs. Helix III and IV are most dissimilar in primary sequence, as observed in other eukaryotic U1 snRNAs (Supplemental Fig. 1A). Helix IV is preceded by a uridine-rich region that may correspond to the Sm-binding domain in T. vaginalis. The primary sequence of U1 helix II apical loop is highly conserved in T. vaginalis, suggesting that role for an U1A protein homolog in the T. vaginalis U1 snRNP. The binding region for U1-70K that was used to isolate the U1 snRNA, the helix I loop, lacks two nucleotides that are invariant in other U1 snRNAs (Yuo and Weiner 1989). There is an apparent deletion of an adenosine (position 2 in the loop of other U1 snRNAs, GAUC > GUC; Supplemental Fig. 1A) that is required for binding of U1-70K to human U1 snRNA (Surowy et al. 1989). Additionally, there is an A > G substitution in the second to last position of this loop.
The T. vaginalis U5 snRNA sequence is also conserved, particularly near the top of helix I (Fig. 1C). Notably, the apical 11-nt loop is identical to that found in the human U5 snRNA. Such conservation throughout eukaryotic evolution likely reflects its essential role in splicing (O'Keefe et al. 1996). This predicted helix I in the T. vaginalis U5 also contains internal loops with the invariant CCG sequence in a loop on the 3′ side of the helix. Helix II, which is preceded by a putative Sm-binding site, has a lower sequence conservation than helix I.
Consistent with splicing mechanisms in other organisms, the T. vaginalis U6 snRNA has the potential to interact with both U2 and U4 snRNAs (Figs. 1B, 2). Notably, the putative U6 snRNA has the predicted invariant sequences (A53CAGAGA59 and A65GC67) found in all examined U6 snRNAs (Figs. 1B, 2A). Sites in U6 snRNA thought to coordinate a metal ion required for catalysis (Yu et al. 1995) are also conserved, and U6/U2 snRNA interaction model reveals three conserved U2–U6 helices and the intramolecular U6 stem–loop ISL I (Fig. 2A). The predicted T. vaginalis U2/U6 snRNA catalytic model is strikingly similar to yeast and human models (Fig. 2A,B), involving two possible conformations (Sun and Manley 1995; Hilliker and Staley 2004).
Putative U2 and U6 snRNAs can efficiently form a complex in vitro
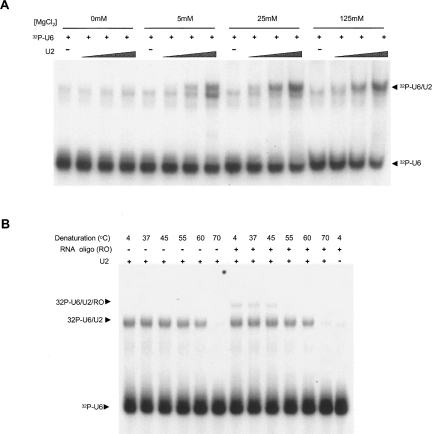
To confirm the identity of putative snRNAs, we tested whether candidate U2 and U6 snRNAs form a stable heterodimer in vitro, as in vitro splicing assays have not been established for T. vaginalis. RNAs corresponding to the central domain of U6 snRNA (nt 41–111) and the 5′ end domain of U2 snRNA (nt 1–47) were assayed for interaction, as demonstrated for the human U2–U6 snRNA complex (Valadkhan and Manley 2000). Initially, the RNAs were mixed in increasing molar ratios, with and without MgCl2, heat denatured, and allowed to return to room temperature. U6 snRNA was 32P-labeled, allowing the formation of a U2–U6 heterodimer to be evaluated by tracking its migration on native polyacrylamide gels. The putative T. vaginalis U2 and U6 RNAs form a MgCl2-dependent complex (Fig. 3A). The formation of this complex increases with increasing concentrations of the unlabeled U2 snRNA and Mg2+, whereas monovalent salts, sodium and ammonium, do not support this U2–U6 snRNA interaction (data not shown). A faint band that migrates slightly faster is also seen in the absence of the U2 oligomer, which is likely the result of weak homodimerization of the U6 oligomer. These data strongly suggest that the identified genes encode bona fide U2 and U6 snRNAs. The predicted structural interaction between U6 snRNA and the putative U4 snRNA, in turn, gives credence to the identity of the U4 gene.
FIGURE 3.
In vitro interaction of T. vaginalis U2/U6 catalytic domains. (A) 32P-labeled U6 RNA (32P-U6) was annealed with increasing concentrations of unlabeled U2 RNA (U2) and Mg2+ as indicated. Lower and higher arrows correspond to free 32P-U6 and a U2–U6 complex, respectively. (B) Interaction of U2–U6 catalytic domains with a RNA oligonucleotide that mimics the intron branch site and evaluation of thermostability of the complexes. 32P-labeled U6 RNA (32P-U6) was annealed with unlabeled U2 RNA (U2) and 20 nM of the RNA oligo (RO), as indicated, in the presence of 20 mM of MgCl2. Annealed complexes were then denatured at the indicated temperatures, and complex formation and stability was assessed by nondenaturing PAGE. Lower, middle, and higher arrows correspond to free 32P-U6, the U2–U6 complex, and the U2–U6-RO complex, respectively.
To further investigate the proposed T. vaginalis U2–U6 interaction, a third RNA (5′-UACUAACACUUA-3′) that mimics the conserved intron branch point sequence and is predicted to interact with U2/U6 snRNA complex (via U2 snRNA nt 28–34, 5′-UGAUGUG-3′) (Fig. 2A) was included in the reaction. Formation of a second, larger complex was dependent on the presence of this unlabeled RNA oligo (Fig. 3B). Denaturation and renaturation experiments further demonstrated that the interaction of the RNA oligo with the U2–U6 complex (Tm between 45°C and 55°C) is less thermostable than the highly stable U2–U6 snRNA interaction (Tm between 60°C and 70°C) (Fig. 3B). Together, these data indicate that T. vaginalis U2 and U6 snRNAs are capable of interacting with each other and with a branch-site motif.
Trichomonas vaginalis snRNAs have uncapped 5′-termini
To gain further insight into the evolution of splicing in this highly divergent eukaryote, we characterized the terminal structures of the identified snRNAs. Spliceosomal U1, U2, U4, and U5 snRNAs typically have 5′-end m2,2,7G (TMG) cap structures (Busch et al. 1982), whereas U6 snRNA contains γ-methyl triphosphate ends (Singh and Reddy 1989). To determine whether T. vaginalis snRNAs have 5′ cap structures, comparative immunoprecipitation of human and T. vaginalis total RNA with a monoclonal antibody that recognizes the TMG cap was performed. Bound and unbound RNA fractions were collected, gel-fractionated, blotted, and hybridized with specific 32P-labeled DNA oligomers. Only negligible amounts of all five T. vaginalis snRNAs bound the anti-TMG antibody (Fig. 4A), whereas the HeLa control U2 and U4 snRNAs were efficiently bound as expected (Speckmann et al. 2000). We also attempted to identify capped RNAs by immunoprecipitating total RNA with the anti-TMG antibody, followed by pCp labeling, and saw no detectable small RNAs using T. vaginalis RNA. In contrast, small RNAs were precipitated and labeled using human RNA as a positive control (data not shown). Cross-reactivity of anti-TMG antibody with the monomethylated caps of mRNAs from T. vaginalis and HeLa cells was observed (Fig. 4A). Together, these data suggest that T. vaginalis snRNAs either have a different cap structure not recognized by the anti-TMG antibody, or lack a 5′ cap. As a control, T. vaginalis and human 5S RNAs were also examined, and neither were recognized by this antibody, indicating that the T. vaginalis 5S RNA likewise lacks a methylated 5′-end cap structure.
To further characterize the 5′ ends of T. vaginalis snRNAs, we utilized RLM-5′RACE. Transcripts known to have a canonical 5′ methylated cap structure (human snRNAs and GAPDH mRNA) as well as those with 5′ monophosphates (T. vaginalis and human 28S rRNAs) were included as controls. Prior to subjecting RNA to reverse transcription (RT), samples were either untreated, treated with calf-intestine alkaline phosphatase (CIP), tobacco acid pyrophosphatase (TAP), or first with CIP and then TAP. An RNA adapter was then ligated to the 5′-end of the RNAs to provide an anchor primer for PCR. CIP digestion will dephosphorylate the 5′ end of an uncapped RNA, making it unavailable for ligation to a RNA adapter. Capped RNAs are protected from digestion by CIP, and require digestion by TAP, to remove the cap, to allow ligation to the adapter. As amplification of a transcript is dependent on the availability of its 5′-end for ligation of the RNA adapter, this method can be used to distinguish whether a target RNA has a protected or unprotected 5′ end. As predicted, the control uncapped 28S RNA transcripts in both human and T. vaginalis were amplified by RLM-5′RACE in the absence of any treatment (Fig. 4B). In contrast, mRNAs from both cell types were amplified only if the RNA was TAP treated, confirming that T. vaginalis mRNAs are capped, as is typical of eukaryotic mRNAs. Amplification of human U2 and U4 snRNAs was also dependent on TAP treatment, as expected. In contrast, the four tested T. vaginalis snRNAs could be amplified by RLM-5′RACE in the absence of any treatment, as observed for the uncapped 28S control transcript, indicating that these RNAs do not have 5′-end cap structures. Furthermore, phosphatase treatment abolished amplification of T. vaginalis snRNAs, confirming that their 5′ ends are uncapped. To exclude the possibility that 5′ end degradation is responsible for the amplification of T. vaginalis snRNAs in the absence of TAP treatment, to confirm the specificity of the PCR product, and to map the precise 5′ end of these RNAs, all products were cloned and sequenced. As shown in Figure 4C, primer extension analyses confirmed the correct 5′ end and the presence of full-length snRNAs for U4, U5, and U6 snRNAs, eliminating the possibility that unprotected 5′ ends are generated by RNA degradation during the procedure. Although 5′-monophosphate ends are required for amplification by the RLM-5′-RACE procedure, endogenous snRNAs in T. vaginalis could contain 5′-triphosphate ends, which are partially converted to 5′-monophosphates either in the cell or during the RNA extraction procedure. Nonetheless, these results demonstrate the lack of a 5′ cap structure on all examined T. vaginalis snRNAs, an unexpected finding, as snRNAs in other examined eukaryotes possess cap structures.
Trichomonas vaginalis snRNAs are transcribed by distinct RNA polymerases
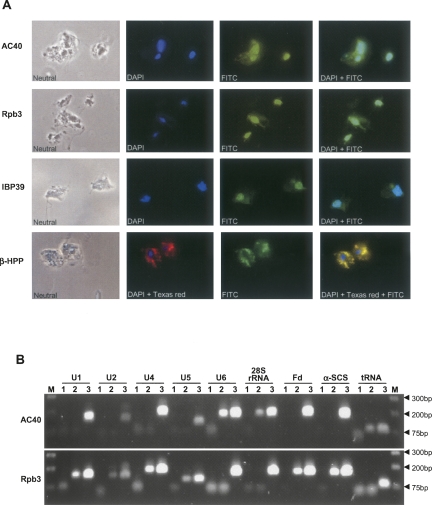
As RNAP II transcripts usually contain either monomethyl (mRNAs) or TMG (snRNAs) caps, the lack of a cap at the 5′ end of T. vaginalis snRNAs raised the question of the identity of RNAP responsible for their transcription. In other organisms, snRNAs are transcribed by RNAP II, except for snRNA U6, which is transcribed by RNAP III. To determine which RNAP transcribes T. vaginalis snRNA genes, we have examined whether subunits specific to RNAP I/III or RNAP II interact with their promoter region and characterized transcription termination signals. To address the subunit composition of the RNAP that transcribes these genes, Rpb3 (Accession no. XP_001313008) and AC40 (Accession no. XP_001317116), core components of RNAP II and I/III, respectively, were used in chromatin immunoprecipitation (ChIP) analyses. A hemaglutinin (HA) tagged was added to the 3′ end of T. vaginalis orthologs of Rpb3 and AC40, and each gene was then individually introduced into T. vaginalis by transfection. HA-tagged IBP39 and β-HPP, nuclear and hydrogenosomal T. vaginalis proteins, respectively (Lau et al. 2003; Brown et al. 2007), were included as controls. Both AC40 and Rpb3 proteins were shown to localize in the nucleus of T. vaginalis by immunofluorescence (Fig. 5A), similar to the IBP39 nuclear control protein and unlike that of the negative hydrogenosomal control protein β-HPP. Interestingly, AC40 localized in a specific nuclear region, while Rpb3 was uniformly distributed throughout the nucleus. The punctuate localization of AC40 is consistent with the clustering of genes transcribed by RNA polymerase I and III (Carlton et al. 2007), consistent with the T. vaginalis AC40 being specific for RNAP I/III complexes. To definitively demonstrate this, ChIP assays of transfectants were performed using the monoclonal anti-HA antibody to immunoprecitate protein/DNA complexes, followed by PCR amplification using primers specific to known RNAP I, II, and III transcripts. These data show that AC40 and Rpb3 specifically coimmunoprecipitate with genes transcribed by RNAP I/III and RNAP II, respectively, confirming their identities as RNAP-specific subunits (Fig. 5B). ChIP analyses with snRNA genes demonstrate that T. vaginalis U1, U2, U4, and U5 genes are associated exclusively with Rpb3, whereas the U6 gene is associated exclusively with AC40. These data indicate that U1, U2, U4, and U5 snRNAs are transcribed by RNAP II, whereas the U6 snRNA is transcribed by RNAP III (Fig. 5B).
FIGURE 5.
Chromatin Immunoprecipitation (ChIP) analysis of RNAP components in T. vaginalis. (A) Localization of HA-tagged RNAP core components AC40 and Rpb3 compared with nuclear HA-tagged IBP-39 (positive control) and hydrogenosomal HA-tagged β-HPP (negative control) proteins. The protein detected by FITC staining is indicated to the left. DAPI was used to stain nuclei; merged FITC and DAPI images are shown at the far right. All images are at a 630X magnification. (B) ChIP analysis of AC40 and Rpb3 T. vaginalis transfectants. Target genes are indicated at the top, Molecular weight markers (M) flank the samples, with molecular weight markers listed in base pairs at the far right. Products of independent ChIP assays to detect AC40 and Rpb3 (top and bottom, respectively) are shown. The following DNA samples were tested by PCR: (1) DNA precipitated from ChIP assay done with wild-type nontransfected T. vaginalis cells (negative control); (2) DNA precipitated from ChIP assay done with transfected HA-tagged AC40 (top) or Rpb3 (bottom); (3) total sheared DNA from wild-type nontransfected cells (PCR positive control).
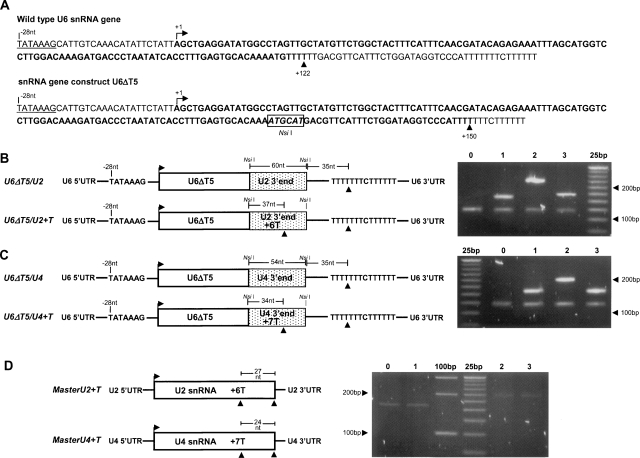
As an independent test of the RNAP(s) that transcribe T. vaginalis snRNA genes, we examined the 5′ and 3′ flanking regions of U2, U4, and U6 snRNAs for putative promoter and terminator elements. The T. vaginalis U6 snRNA gene contains a TATA-box at –28 nt and a cluster of six T residues immediately downstream from its 3′ end (Fig. 6A). TATA-box elements are known to facilitate initiation of transcription by RNAP II and III, whereas a cluster of four or more T residues is specifically involved in transcription termination by RNAP III. To test whether this T-stretch is required for termination of transcription of the T. vaginalis U6 gene, T. vaginalis transfectants harboring a construct (U6ΔT5) where this T-stretch was mutated (ATGTTTTT to CATGAC) were made, and transcription termination of the gene was tested. The 3′ end of the U6 gene also contains a T-stretch ∼30 nt downstream from the mutated region, which was not altered in this construct (Fig. 6A). The 3′ RACE analyses demonstrated that in addition to the endogenous U6 snRNA transcript, U6ΔT5 transfectants produce an additional ∼30 nt longer U6 transcript, which corresponds to termination at the downstream T-stretch (Fig. 6B,C), as confirmed by sequencing. These results show that termination of U6 gene transcription in T. vaginalis is marked by a stretch of Ts, a characteristic specific for RNAP III transcription.
FIGURE 6.
Evaluation of snRNA transcription termination by 3′ RACE. (A) Sequence of endogenous and mutated U6 snRNA genes. A putative TATA-box at −28 nt is underlined, and mapped transcription units are in bold. Both transcription units terminate at a cluster of T residues. (B) 3′ RACE analysis of U6 snRNA from wild-type nontransfected T. vaginalis cells (0) and from T. vaginalis cells transfected with the plasmid constructs: U6ΔT5 (1), U6ΔT5/U2 (2), U6ΔT5/U2+T (3). (C) 3′RACE analysis of U6 snRNA from wild-type nontransfected T. vaginalis cells (0) and T. vaginalis cells transfected with plasmid constructs: U6ΔT5 (1), U6ΔT5/U4 (2), U6ΔT5/U4+T (3). (D) 3′ RACE analysis of U2 snRNA (0 and 1) and U4 snRNA (2 and 3) from wild-type nontransfected T. vaginalis cells (0 and 2) and from T. vaginalis cells transfected with plasmid constructs: MasterU2+T transfectant (1) and MasterU4+T transfectant (3).
As not only a cluster of T residues but also the surrounding nucleotides have been observed to affect RNA polymerase III termination (Braglia et al. 2005), we tested whether clusters of T residues in the context of U2 and U4 snRNA 3′ end sequences would serve as terminators for U6 gene transcription (Fig. 6B,C). Sequences surrounding the 3′ end of U2 and U4 snRNAs were inserted at the 3′-end of the U6ΔT5 construct, generating plasmids carrying U6/U2 and U6/U4 chimeras (plasmids U6ΔT5/U2 and U6ΔT5/U4). Mutations were also introduced in these constructs to generate a cluster of T residues in the inserted U2 and U4 snRNA 3′ sequences (plasmids U6ΔT5/U2+T and U6ΔT5/U4+T) (Fig. 6B,C). All constructs were transfected into T. vaginalis and 3′ RACE and DNA sequencing were used to evaluate transcription termination. In this context, the inserted T cluster was also recognized as a terminator. To directly examine the termination of transcription of endogeneous U2 or U4 snRNA transcription, the same T cluster was inserted into constructs containing U2 or U4 genes with their endogeneous upstream and downstream sequences (Fig. 6D). Mapping of the 3′ end of the resulting transcripts, by sequencing cloned RT–PCR products, illustrates that U2 and U4 gene transcription is not terminated by this T stretch, and instead, termination occurred at the same 3′-end nucleotide as the endogenous genes. These findings confirm the ChIP data indicating that the same RNAP transcribes U2 and U4 genes and a different one transcribes the U6 gene. Taken together, these data indicate that T. vaginalis U6 gene transcribed by RNAP III and T. vaginalis U2 and U4 genes are transcribed by RNAP II.
DISCUSSION
Relatively few genes (∼65) in the T. vaginalis genome appear to contain an intron (Carlton et al. 2007). The introns that have been identified are uniformly short and characterized by a conserved 12-nt sequence (5′-ACTAACACACAG/-3′) that contains both the branch site (underlined) and 3′ splice site (/), with the former separated by only 7 nt from the latter (Vanacova et al. 2005; Carlton et al. 2007). Unusually short introns and the fusion of the branch site and 3′ splice site are also found in the four known introns in G. lamblia, another highly divergent eukaryote (Morrison et al. 2007). Prior to this study, spliceosomal snRNAs required for splicing catalysis in other eukaryotes had not been identified from either Trichomonas or Giardia. Here we have identified all five T. vaginalis spliceosomal snRNAs. These snRNAs can be folded into evolutionarily conserved secondary structures and contain sequences and structural motifs known to be required for splicing activity in other organisms (Guthrie and Patterson 1988). Sm-binding motifs, necessary for binding the core components of spliceosomal snRNPs, were found at conserved positions in all T. vaginalis snRNAs (Fig. 1). The consensus of these motifs, AAU3NR3, is similar to the established consensus RAU5GR, in contrast to the degenerate sequence found in kinetoplastids (Mottram et al. 1989; Hinas et al. 2006). Once snRNAs are identified in Giardia, should they be found to also be conserved, it would be tempting to propose that the core structure of modern spliceosomes was established in basal eukaryotes. Should this be the case, the kinetoplastids would appear to have diverged from this basal core structure.
The high degree of conservation observed for T. vaginalis spliceosomal snRNAs suggests that not only the mechanism of splicing, but also most steps involved in intron recognition and rearrangement of spliceosomal RNA structure have been conserved throughout eukaryote evolution. These include the formation of commitment and pre-spliceosome complexes. Both primary sequence and secondary structure of T. vaginalis U1 snRNA exhibit a high degree of conservation with other eukaryotes. Conservation of its 5′ end indicates an invariant mechanism for recognition of the 5′SS. In addition, the presence of a highly conserved apical loop at U1 snRNA helix II suggests conservation of interacting U1-specific splicing factors. Furthermore, the T. vaginalis U1 snRNA was isolated based on its interaction with a T. vaginalis U1-70k homolog, known to interact with the helix I loop of other eukaryotic U1 snRNAs (Surowy et al. 1989; Yuo and Weiner 1989). Despite the conservation of this interaction in T. vaginalis, its U1-70 k homolog (GenBank XP_001303000) is significantly smaller (∼31 kDa) than its counterparts in other eukaryotes (Yuo and Weiner 1989). Moreover, the conserved region in helix I (GAUC) (Surowy et al. 1989), to which it is predicted to bind, has a nucleotide deletion (GUC).
U2 snRNA also binds to pre-mRNA within the pre-spliceosomal complex and undergoes changes in secondary structure during spliceosome rearrangements and splicing catalysis (Hilliker et al. 2007; Perriman and Ares Jr. 2007). Our structural prediction models strongly support the ability of the T. vaginalis U2 snRNA to adopt secondary structures and to interact with U6 snRNA and the conserved intron branch-site motif as previously described (Ares Jr. and Weiser 1995). The T. vaginalis U2 snRNA contains the invariant branch-site interaction sequence 5′-GUAGUA-3′ that can stably base pair to the unusually conserved branch site found in all T. vaginalis introns (Vanacova et al. 2005). T. vaginalis introns lack polypyrimidine tracts between the branch site and 3′SS, and thus are unlikely to bind U2 auxiliary factors (U2AFs) known to stabilize the branch site–U2 snRNA interaction in metazoa (Ares and Weiser 1995), suggesting that such factors may not be required/present in the T. vaginalis spliceosome.
We have demonstrated that T. vaginalis U2 and U6 snRNAs form stable complexes in vitro (Fig. 3). Sequence analysis allows for building models of base-pairing interactions between U6/U2 and U6/U4 snRNAs. By analogy to snRNAs in other organisms, these models accommodate conformational changes thought to take place during the spliceosome assembly leading to the formation of a catalytically active spliceosome (Nilsen 1998). Two strictly conserved U6 sequence motifs, ACAGAGA and AGC, are implicated in formation of the catalytic center and its rearrangements (Nilsen 1998). The strong similarity of T. vaginalis U2/U6 snRNA interaction model to that of other examined eukaryotes underscores the phylogenetic conservation of critical interactions (Fig. 2) that allow the formation of two mutually exclusive conformations proposed to facilitate the two-step splicing catalysis (Sashital et al. 2004). Furthermore, similar U2/U6 snRNA structure has been argued to be involved in an in vitro protein-free reaction that resembles certain aspects of splicing (Valadkhan and Manley 2001, 2003; Sashital et al. 2004). As discussed above, our model also gives support to the phylogenetic conservation of U2/U6 helix I, divided by a 2-nt bulge into helix Ia and Ib (Madhani and Guthrie 1992; McPheeters and Abelson 1992). In yeast, genetic evidence supports the formation of a tertiary interaction mediated by a bulged U2 adenosine (A25) found between helix Ia and Ib and the guanosine G52 found in the U6 evolutionary conserved sequence A47CAGAGA53 (yeast numbering) (Madhani and Guthrie 1994). Although we do not have evidence of such tertiary interaction, conservation of helix Ia and Ib in T. vaginalis U2/U6 interactions further supports a role for helix I in splicing (Fig. 2).
T. vaginalis U6 intramolecular stem–loop (ISL) appears to be less stable than its counterparts in yeast or metazoa. However, genetic analyses in yeast indicate that the stability of U6-ISL is not as important as its sequence (McPheeters 1996) and that it is possible that other RNA structures or proteins that interact with U6-ISL compensate for its apparent destabilization in T. vaginalis. The lack of stable U6-AGC base pairing to the U6-ISL in T. vaginalis might indicate folding instability of this structure. Nevertheless, our model supports the importance of the upper portion of this stem (McPheeters 1996; Shukla and Padgett 2001) as well as the formation of the U6 ISL pentaloop GNR(N)A, similar to that proposed in other organisms (Huppler et al. 2002). Structure-function analyses of the T. vaginalis spliceosomal proteins and snRNAs will be necessary to test whether these structural differences impact splicing mechanisms in this divergent eukaryote. It is possible that this and other differences in stabilities of various RNA–RNA interactions within snRNAs in different organisms reflect modulation of relative stabilities of spliceosomal conformations along the splicing pathway. The U6 ISL sequence also affects U4–U6 pairing, which in T. vaginalis represents a stable, perfect Watson-Crick interaction, whereas human U4–U6 stem II contains several U-G pairs. At present, our understanding of such structural rearrangements is insufficient to predict detailed consequences of the observed sequence differences between T. vaginalis and metazoan snRNAs.
In addition to the sequence and structural similarities of the newly identified T. vaginalis spliceosomal snRNAs compared with those in yeast and metazoa, a high degree of conservation in another important component of the spliceosome, Prp8p, has been previously described (Fast and Doolittle 1999). This large (280 kDa) component of U5 snRNP is involved in multiple interactions near the catalytic center (Grainger and Beggs 2005) and implicated in modulation of spliceosomal transitions between the catalytic steps (Liu et al. 2007). The T. vaginalis PRP8 is also highly conserved; over its entire length it shares 56% amino acid identity with the human and Caenorhabditis elegans proteins. These observations further support our argument based on structural similarities of snRNAs that the splicing apparatus and splicing mechanisms have been highly conserved among all eukaryotes.
An unusual property of T. vaginalis snRNAs, however, is the absence of a characteristic 5′-terminal TMG cap, found in other organisms (Mouaikel et al. 2002). Instead, we found that T. vaginalis snRNAs have unmodified 5′ termini. The biological significance of a TMG 5′-cap structure on snRNAs is not fully understood; however, roles in trafficking and snRNP assembly have been proposed (Huber et al. 2002; Mouaikel et al. 2002; Narayanan et al. 2003; Cougot et al. 2004). Whether the lack of a 5′ cap influences trafficking and/or assembly of T. vaginalis snRNAs awaits further analyses. It is notable that although S. cerevisiae and Schizosaccharomyces pombe snRNAs have TMG caps, its presence is not essential, as deletion of the enzyme required for formation of a TMG cap, trimethylguanosine synthase (Tgs), results in snRNAs lacking TMG caps, but does not affect viability (Mouaikel et al. 2002; Hausmann et al. 2007). On the other hand, similar studies in Drosophila have shown that the absence of TMG cap synthesis and TMG-containing RNAs result in embryonic lethality (Komonyi et al. 2005).
As capping of transcripts is coordinated with transcription and is specific for different RNAPs, we determined which RNAP(s) transcribe T. vaginalis snRNAs. These data indicate that U1, U2, U4, and U5 genes are transcribed by RNAP II, despite the absence of a 5′ cap structure typically formed on nascent transcripts early during RNAP II transcription (Cougot et al. 2004). Using the same experimental approaches, T. vaginalis mRNAs, also transcribed by RNAP II (Liston et al. 2001; Schumacher et al. 2003), were found to contain a 5′ cap structure. Thus, the absence of a cap on the snRNA transcripts does not reflect a general lack of capping of T. vaginalis RNAP II transcripts. Whether T. vaginalis RNAP II snRNA transcripts are transcribed by a variant RNAP II complex that does not associate with fully active capping machinery or they originally contain a cap that is subsequently removed by 5′ end processing, is unknown.
T. vaginalis U6 snRNA was found to be transcribed by RNAP III, consistent with structural properties of the gene. This U6 gene lacks an A/B box and has a TATA-box at position −28 nt similar to the mammalian RNAP III U6 snRNA genes that rely on a extragenic 5′ promoter (Hernandez 2001). The 3′ end of the T. vaginalis U6 snRNA is also flanked by a cluster of T-residues, which was demonstrated to mark termination of transcription, a characteristic of RNAP III transcripts.
Together, our analyses illustrate that T. vaginalis snRNAs are unique among their eukaryotic counterparts, based on the absence of a 5′-end cap structure, but are otherwise extremely conserved based on predicted secondary structure models, indicating a strong conservation of splicing mechanisms throughout eukaryotic evolution. Availability of these snRNAs sets the stage for the isolation and characterization of spliceosomes from this divergent eukaryote, allowing further analyses of the conservation of core spliceosomal proteins in diverse eukaryotes.
MATERIALS AND METHODS
Identification of T. vaginalis snRNAs
The 7× coverage of the T. vaginalis genome sequence database (http://www.tigr.org/tdb/e2k1/tvg/) was downloaded to create a local database that was searched using BioEdit software (matrix PAM40 setting) to identify trichomonad snRNAs. Candidate U2, U4, and U5 snRNAs were identified initially using a BLASTn search, followed by structural analyses (described below); U6 and U1 snRNA identification required a more involved bioinformatics approach and functional assays, respectively. Three candidate U2 genes resulted from a BLASTn search, using CAAGTGTAGTATCTG (conserved nucleotides 28–42 in human U2 snRNA). RNAfold software (Hofacker 2003) was then used to generate predicted structures from these three sequences and their upstream and downstream regions. A subsequent search for conserved nucleotides and predicted U2 helix I and IIa lead to the identification of a single U2 snRNA gene. A BLASTn search, using TTCGCCTTTTACTA (conserved nucleotides 34–n47 in human U5 snRNA) likewise resulted in three candidate U5 genes that were then subjected to the structural analyses. A single U5 snRNA gene, capable of forming the stem–loop I with its 3′ conserved CCG, was identified. The U6 snRNA was identified using INFERNAL software (ftp://ftp.genetics.wustl.edu/pub/eddy/software/infernal), snRNA databases, and covariance models from The Sanger Institute Rfam database (http://www.sanger.ac.uk/Software/Rfam/index.shtml). A multiple U6 snRNA alignment was built and used to screen the 7X T. vaginalis genome database using the default parameters of the cmsearch program. The resulting output revealed three hits; however, only the top hit had a significant score value. Further examination of this hit showed it to have the two U6 snRNA invariant sequences ACAGAGA and AGC, whereas the other two did not. To identify U4 snRNA, a BLASTn search using U6 snRNA nucleotides 71–85 (CTCCTTGGACAAGA) predicted to interact with U4 was done, resulting in 12 candidate genes. Each sequence was searched for a possible intramolecular stem–loop in nucleotide positions 30–35, followed by 10–15 nt that could interact with the candidate U6 snRNA to form a Y-shaped U4–U6 interaction domain. This approach resulted in the identification of a single U4 snRNA gene.
U1 snRNA was not successfully identified using bioinformatics. To isolate U1 snRNA, the T. vaginalis homolog of U1-70K (GenBank accession number XM_001302999.1) was tagged with streptoavidin binding protein (SBP) at its C terminus and cloned into the Master-Neo plasmid (Dyall et al. 2003). Nuclear extracts of T. vaginalis cells transfected with the resulting construct were prepared as described (Mayeda and Krainer 1999), except parasites were lysed by passage through a 25G5/8 needle syringe. Approximately 0.6 μg of nuclear protein extract was mixed with 300 μL of prewashed streptavidin Sepharose (Amersham) in a final volume of 1.2 mL of 20 mM Hepes (pH7.9); 70 mM KCl, 20 mM MgCl2, 2 mM EDTA, 0.05% NP-40, 0.5 mM DTT, plus protease inhibitors (Roche complete mini, EDTA-free, cocktail). After 1 h incubation at 4°C, the Sepharose was washed 8X with 1 mL of the same buffer, and RNA was extracted by Trizol LS extraction (Invitrogen). [32P]pCp labeling of the recovered RNA was performed and visualized in a TBE-Urea 5% polyacrylamide gel by autoradiography. RNAs ranging in size from 100 to 200 nt were recovered from the gel, polyadenylated with recombinant Escherichia coli Poly-A Polymerase I (Ambion), phenol extracted, ethanol precipitated, and reverse transcribed using a polydT 2-base anchor primer adapter, as described (FirstChoice RLM-RACE kit, Ambion). cDNA was then isolated by gel electrophoresis and poly-C tailed at the 5′ terminus by terminal deoxynucleotidyl transferase. Outer and inner PCR products were obtained by using GI-rich adapter primers (5′ RACE system, Invitrogen) and specific primers for the polydT adapter (FirstChoice RLM-RACE kit, Ambion). All PCR products were cloned into the Topo pCR-4 vector (Invitrogen) and sequenced using M13 primers and the Big Dye version 3 labeling kit (Applied Biosystems).
In vitro interaction of U2–U6 minimal catalytic domains
The region corresponding to the central domain of U6 snRNA (nt 41–111) was transcribed in a large scale T7-transcription system (Ambion) and oligonucleotides corresponding to the 5′ domain of U2 snRNA (nt 1–47) and RNA branch oligonucleotide (RO; UACUAACACUUA) were purchased from Invitrogen. Following acrylamide gel purification of the RNAs, an in vitro interaction assay was performed as described previously (Valadkhan and Manley 2000). U2 or U6 RNA domains labeled at the 3′-end with [32P]pCp and RNA ligase (Ambion) were incubated in the presence of 5 nM of the unlabeled U2 snRNA and 0, 1, 5, and 10 nM of the 32P-labeled U6 snRNA, and vice versa, in the presence of 0, 5, 25, and 125 mM of MgCl2. Reactions, including the RO, contained 5 nM of 32P-labeled U6 RNA, 10 nM of the unlabeled U2 RNA (U2), 20 nM of the RO), and 20 mM of MgCl2. Complexes were then treated for 15 min at 4, 37, 45, 55, 60, or 70°C before loading on a polyacrylamide gel and analyzed by gel electrophoresis as described (Valadkhan and Manley 2000).
T. vaginalis culture and transfections
T. vaginalis strain T1 was maintained in TYM medium supplemented with 10% horse serum and iron (Clark and Diamond 2002). Transfections were conducted as described (Delgadillo et al. 1997), and cultures were selected with Gentamicin (GIBCO). Within 10 d of cell culturing, DNA, RNA, or protein preparations were prepared from transfectants.
Plasmid constructs, chimeras, and mutagenesis
All gene constructs used the Master-Neo plasmid, a T. vaginalis expression vector for stable transfection that contains the neomycin resistance gene for selection and NdeI and Asp718 sites for cloning genes of interest immediately upstream of a double-HA epitope tag (Dyall et al. 2003). Constructs of genes containing native 5′ UTR and/or 3′ UTRs were made by digesting MasterNeo, followed by ligation with PCR-generated Rpb3, U2, U4, or U6 genes. Site-directed mutagenesis was performed by PCR, and the mutation was confirmed by sequencing. U6–U2 and U6–U4 chimeras were made by inserting a NsiI restriction site to delete the U6 snRNA terminator. The last 37 and 34 nt of U2 and U4 snRNA sequences, respectively, were inserted into the 3′ end of U6 snRNA to produce the chimeras. In U2 snRNA, positions 147 and 149 were mutated to generate a stretch of six T residues, as underlined (146TGTCTTCAC154 to 146TTTTTTCAC154). In U4 snRNA, positions 124 and 125 were mutated to generate a stretch of seven T residues, as underlined (121CTTAATTTC129 to 121CTTTTTTTC129).
RNA isolation and analysis
Total RNA was isolated by mirVana™ PARIS™ isolation kit (Ambion). For snRNA analyses, RNA was separated on either TBE-Urea 5% acrylamide or formaldehyde agarose gels. RNA gels were transferred to nylon membranes using standard procedures (GE Healthcare) and hybridized with 32P-labeled probes using recommended stringency (Ambion). For the snRNAs, hybridizations were confirmed using three independent DNA oligomers as probes.
Immunoprecipitation of RNA by antitrimethylguanosine (TMG) monoclonal antibody
Twenty microliters of prewashed anti-2,2,7-trimethylguanosine (TMG) antibody agarose beads (Calbiochem), were incubated with 100 μg of total RNA in 0.5 mL 01.% Triton X-100, 50 mM NaCl, and 20 mM HEPES (pH 7.9) for 1 h at 4°C. The supernatant was recovered as the unbound RNA and precipitated with ethanol. Beads were washed eight times with 0.5 mL incubation buffer by repeated centrifugation. Elution was then performed twice by incubating beads with 100 μL of 0.5% SDS, 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 10 μg/mL of proteinase K at 55°C for 15 min each. Recovered bound RNA was phenol/chloroform extracted and ethanol precipitated.
Mapping of 3′ and 5′-ends of snRNAs by primer extension and DNA sequencing
RNA ligase-mediated rapid amplification of cDNA 5′-Ends (RLM-5′ RACE) was performed with DNAse treated (Ambion) RNA from T. vaginalis and HeLa cells, following recommendations (Invitrogen). From the same RNA sample, different treatments were performed prior to RNA oligomer ligation and reverse transcription, as follows: (1) no treatment, (2) calf-intestine alkalyne phosphatase (CIP) only, (3) tobacco acid pyrophosphatase (TAP) only, (4) CIP followed by TAP treatment. The treated RNA samples were then split in equal parts for RT–PCR reactions using RNA adapter specific primers and gene-specific primers for detection of the following transcript targets: (1) T. vaginalis ferredoxin mRNA and human GAPDH mRNA, (2) T. vaginalis and human 28S RNAs, (3) T. vaginalis and human snRNAs U2, U4, and U6. Results were reproduced in an independent duplicate experiment. All RT–PCR products were cloned and sequenced. For T. vaginalis snRNAs, manual DNA sequencing (SequiTherm Excel II, Epicentre) was performed in parallel to primer extension reactions using the respective gene-specific reverse 32P-labeled primer used in the RLM-5′ RACE. T. vaginalis total RNA was subjected to polyadenylation and 3′ ends of the snRNAs were mapped using 3′ RACE as recommended (Poly(A) tailing kit and FirstChoice RLM-RACE, Ambion).
Immunofluorescence microscopy
T. vaginalis cells were washed with warm phosphate-buffered saline (PBS) and allowed to attach to coverslips for 1–2 h in a humidifying chamber at 37°C. All procedures thereafter were done at room temperature. The cells were fixed with 3.5% formalin in PBS for 20 min, quenched with 100 mM glycine in PBS for 5 min, blocked with 3% BSA in PBS for 15 min, and permeabilized with 3% BSA, 0.2% Triton X-100 in PBS for 15 min. Rpb3 and AC40 HA-tagged transfectants were visualized using mouse anti-HA monoclonal antibody (Covance) and secondary Alexa Fluor 488 (FITC) donkey antimouse antibodies (Invitrogen). HA-tagged IBP39 (Lau et al. 2003) and β-HPP (Brown et al. 2007) transfectants were localized as previously described. For each antibody, 1 h incubation was followed by five PBS washes. Coverslips were then mounted onto slides using ProLong Gold with DAPI (Invitrogen). An Axioscop2 microscope outfitted with Axiovision v. 3.2 software was used to visualize and process images (Zeiss).
Chromatin immunoprecipitation (ChIP) analysis
T. vaginalis transfectants expressing AC40 (GenBank™ accession no. XP_001317116) or Rpb3 (GenBank accession no. XP_001313008) tagged with double-HA epitope at the C terminus were used for ChIP assays as previously described (Ong et al. 2006), except washes were conducted twice with each of the following buffers: (1) 1% Triton X-100, 1 mM EDTA, 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.1% deoxycholate; (2) 1% Triton X-100, 1 mM EDTA, 50 mM HEPES (pH 7.5), 500 mM NaCl, 0.1% deoxycholate; (3) 0.5% NP-40, 1 mM EDTA, 10 mM Tris-HCl (pH 8.0), 250 mM LiCl, 0.5% deoxycholate. For negative controls, either wild-type cells were included or the primary anti-HA antibody was omitted for the transfected cell samples. Sheared DNA from wild-type cells before going to ChIP was used as a positive PCR control. Different sets of primers were used to detect the following transcription targets; (1) 28S rRNA for a RNAP I gene; (2) ferredoxin or α-SCS for RNAP II genes; (3) tRNA for RNAP III genes; (4) U2, U4, and U6 snRNA genes. Forward and reverse primers were designed to detect a product about 150–200 nt, having the forward primer to anneal about 100 nt upstream of the transcription unit. The sole exception was the tRNA gene that primers were designed to detect the full transcript. PCR products were resolved in 1.5% TBE-agarose gels. The experiment was performed in duplicate for each AC40 and Rpb3 transfectants.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Maria Delgadillo-Correa and Camila L. Correa for technical assistance, Alias J. Smith for help with the ChIP analyses, and Drs. Cheryl Okumura, Chris Ryan, and Nancy Sturm for critical comments on the manuscript and our colleagues in the lab for helpful discussions. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant AI30537 to P.J.J., NIH grant GM49044 to M.M.K., and a fellowship to A.S.B. from CNPq-Brazil (PDE 200065/2004-1).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1045408.
REFERENCES
- Abrahamsen M.S., Templeton T.J., Enomoto S., Abrahante J.E., Zhu G., Lancto C.A., Deng M., Liu C., Widmer G., Tzipori S., et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum . Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Ares M., Jr, Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol. 1995;50:131–159. doi: 10.1016/s0079-6603(08)60813-2. [DOI] [PubMed] [Google Scholar]
- Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Braglia P., Percudani R., Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- Brow D.A., Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Brown M.T., Goldstone H.M., Bastida-Corcuera F., Delgadillo-Correa M.G., McArthur A.G., Johnson P.J. A functionally divergent hydrogenosomal peptidase with protomitochondrial ancestry. Mol. Microbiol. 2007;64:1154–1163. doi: 10.1111/j.1365-2958.2007.05719.x. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Thomas T., Sharp P.A. Splicing of precursors to mRNAs by the Spliceosome. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 1999. [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y.C. SnRNAs, SnRNPs, and RNA processing. Annu. Rev. Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Carlton J.M., Hirt R.P., Silva J.C., Delcher A.L., Schatz M., Zhao Q., Wortman J.R., Bidwell S.L., Alsmark U.C., Besteiro S., et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis . Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C.G., Diamond L.S. Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 2002;15:329–341. doi: 10.1128/CMR.15.3.329-341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., van Dijk E., Babajko S., Seraphin B. ‘Cap-tabolism’. Trends Biochem. Sci. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Davis C.A., Brown M.P., Singh U. Functional characterization of spliceosomal introns and identification of U2, U4, and U5 snRNAs in the deep-branching eukaryote Entamoeba histolytica . Eukaryot. Cell. 2007;6:940–948. doi: 10.1128/EC.00059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgadillo M.G., Liston D.R., Niazi K., Johnson P.J. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis . Proc. Natl. Acad. Sci. 1997;94:4716–4720. doi: 10.1073/pnas.94.9.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall S.D., Lester D.C., Schneider R.E., Delgadillo-Correa M.G., Plumper E., Martinez A., Koehler C.M., Johnson P.J. Trichomonas vaginalis Hmp35, a putative pore-forming hydrogenosomal membrane protein, can form a complex in yeast mitochondria. J. Biol. Chem. 2003;278:30548–30561. doi: 10.1074/jbc.M304032200. [DOI] [PubMed] [Google Scholar]
- Ebel C., Frantz C., Paulus F., Imbault P. Trans-splicing and cis-splicing in the colourless Euglenoid, Entosiphon sulcatum . Curr. Genet. 1999;35:542–550. doi: 10.1007/s002940050451. [DOI] [PubMed] [Google Scholar]
- El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N., Ghedin E., Worthey E.A., Delcher A.L., Blandin G., et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Abelson J. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 1992;20:3659–3664. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantoni A., Dare A.O., Tschudi C. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol. Cell. Biol. 1994;14:2021–2028. doi: 10.1128/mcb.14.3.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast N.M., Doolittle W.F. Trichomonas vaginalis possesses a gene encoding the essential spliceosomal component, PRP8. Mol. Biochem. Parasitol. 1999;99:275–278. doi: 10.1016/s0166-6851(99)00017-1. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger R.J., Beggs J.D. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu. Rev. Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Ramirez A., Schneider S., Schwer B., Shuman S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe . Nucleic Acids Res. 2007;35:1411–1420. doi: 10.1093/nar/gkl1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: A model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Hilliker A.K., Staley J.P. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A.K., Mefford M.A., Staley J.P. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinas A., Larsson P., Avesson L., Kirsebom L.A., Virtanen A., Soderbom F. Identification of the major spliceosomal RNAs in Dictyostelium discoideum reveals developmentally regulated U2 variants and polyadenylated snRNAs. Eukaryot. Cell. 2006;5:924–934. doi: 10.1128/EC.00065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J., Dickmanns A., Luhrmann R. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler A., Nikstad L.J., Allmann A.M., Brow D.A., Butcher S.E. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- Komonyi O., Papai G., Enunlu I., Muratoglu S., Pankotai T., Kopitova D., Maroy P., Udvardy A., Boros I. DTL, the Drosophila homolog of PIMT/Tgs1 nuclear receptor coactivator-interacting protein/RNA methyltransferase, has an essential role in development. J. Biol. Chem. 2005;280:12397–12404. doi: 10.1074/jbc.M409251200. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Rymond B.C., Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987;50:593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Lau A.O., Liston D.R., Vanacova S., Johnson P.J. Trichomonas vaginalis initiator binding protein, IBP39, contains a novel DNA binding motif. Mol. Biochem. Parasitol. 2003;130:167–171. doi: 10.1016/s0166-6851(03)00172-5. [DOI] [PubMed] [Google Scholar]
- Liston D.R., Lau A.O., Ortiz D., Smale S.T., Johnson P.J. Initiator recognition in a primitive eukaryote: IBP39, an initiator-binding protein from Trichomonas vaginalis . Mol. Cell. Biol. 2001;21:7872–7882. doi: 10.1128/MCB.21.22.7872-7882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Query C.C., Konarska M.M. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat. Struct. Mol. Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- Loftus B., Anderson I., Davies R., Alsmark U.C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R.P., Mann B.J., et al. The genome of the protist parasite Entamoeba histolytica . Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes & Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Krainer A.R. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- McPheeters D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- McPheeters D.S., Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992;71:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- Morrison H.G., McArthur A.G., Gillin F.D., Aley S.B., Adam R.D., Olsen G.J., Best A.A., Cande W.Z., Chen F., Cipriano M.J., et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia . Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Mottram J., Perry K.L., Lizardi P.M., Luhrmann R., Agabian N., Nelson R.G. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: Identification of the U2, U4, and U6 RNA analogs. Mol. Cell. Biol. 1989;9:1212–1223. doi: 10.1128/mcb.9.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- Narayanan A., Eifert J., Marfatia K.A., Macara I.G., Corbett A.H., Terns R.M., Terns M.P. Nuclear RanGTP is not required for targeting small nucleolar RNAs to the nucleolus. J. Cell Sci. 2003;116:177–186. doi: 10.1242/jcs.00176. [DOI] [PubMed] [Google Scholar]
- Nelissen R.L., Will C.L., van Venrooij W.J., Luhrmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. RNA–RNA interactions in nuclear pre-mRNA splicing. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. [Google Scholar]
- Nilsen T.W. The spliceosome: The most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Nixon J.E., Wang A., Morrison H.G., McArthur A.G., Sogin M.L., Loftus B.J., Samuelson J. A spliceosomal intron in Giardia lamblia . Proc. Natl. Acad. Sci. 2002;99:3701–3705. doi: 10.1073/pnas.042700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe R.T., Norman C., Newman A.J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Ong S.J., Hsu H.M., Liu H.W., Chu C.H., Tai J.H. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis . Eukaryot. Cell. 2006;5:391–399. doi: 10.1128/EC.5.2.391-399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R.J., Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes & Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital D.G., Cornilescu G., McManus C.J., Brow D.A., Butcher S.E. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Lau A.O., Johnson P.J. Structural basis of core promoter recognition in a primitive eukaryote. Cell. 2003;115:413–424. doi: 10.1016/s0092-8674(03)00887-0. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Shukla G.C., Padgett R.A. The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA. 2001;7:94–105. doi: 10.1017/s1355838201000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Reddy R. γ-monomethyl phosphate: A cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl. Acad. Sci. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann W.A., Terns R.M., Terns M.P. The box C/D motif directs snoRNA 5′-cap hypermethylation. Nucleic Acids Res. 2000;28:4467–4473. doi: 10.1093/nar/28.22.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.S., Manley J.L. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Surowy C.S., van Santen V.L., Scheib-Wixted S.M., Spritz R.A. Direct, sequence-specific binding of the human U1-70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol. Cell. Biol. 1989;9:4179–4186. doi: 10.1128/mcb.9.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Valadkhan S., Manley J.L. A tertiary interaction detected in a human U2-U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA. 2000;6:206–219. doi: 10.1017/s1355838200992197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S., Manley J.L. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- Valadkhan S., Manley J.L. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Yan W., Carlton J.M., Johnson P.J. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis . Proc. Natl. Acad. Sci. 2005;102:4430–4435. doi: 10.1073/pnas.0407500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.A., Manley J.L. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991;352:818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- Yean S.L., Wuenschell G., Termini J., Lin R.J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.T., Maroney P.A., Darzynkiwicz E., Nilsen T.W. U6 snRNA function in nuclear pre-mRNA splicing: A phosphorothioate interference analysis of the U6 phosphate backbone. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- Yuo C.Y., Weiner A.M. Genetic analysis of the role of human U1 snRNA in mRNA splicing: I. Effect of mutations in the highly conserved stem-loop I of U1. Genes & Dev. 1989;3:697–707. doi: 10.1101/gad.3.5.697. [DOI] [PubMed] [Google Scholar]