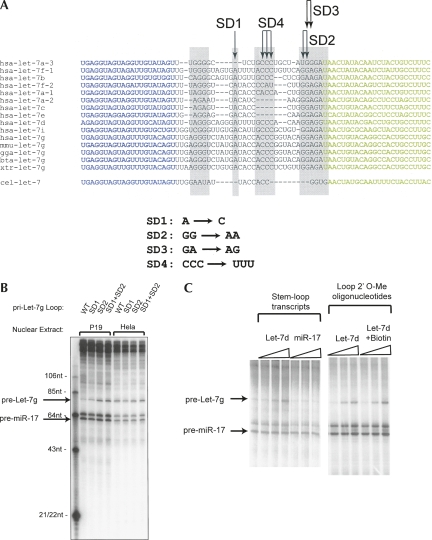
Figure 2.
The loop region of Let-7 interacts with the Drosha Inhibitor. (A) The sequence alignment of Let-7 family members is shown. (Blue text) Mature miRNA sequence; (green text) the complementary stem strand (not exactly the star strand); (gray boxes) regions of homology; (arrows) nucleotides that were mutagenized. Sequence changes are indicated below the alignment. (B) Wild-type (WT) or mutant pri-Let-7g substrates were combined with the pri-miR-17 substrate and incubated in a P19 or HeLa nuclear extract. Drosha products were resolved and are indicated. (C) Pri-miRNA substrates for Let-7g and miR-17 were combined and incubated in P19 nuclear extracts. Competitor RNA transcripts corresponding to the loop plus 12 nt of each stem, or competitor 2′-O-methyl oligonucleotides, were included at 10, 50, and 250 nM final concentration. In one case, the oligonucleotide had a 3′ Biotin moiety. The left lane had no competitor. Drosha products were resolved on a denaturing polyacrylamide gel. (Arrows) Precursor products.